Transcriptome classification reveals molecular subtypes in psoriasis
- PMID: 22971201
- PMCID: PMC3481433
- DOI: 10.1186/1471-2164-13-472
Transcriptome classification reveals molecular subtypes in psoriasis
Abstract
Background: Psoriasis is an immune-mediated disease characterised by chronically elevated pro-inflammatory cytokine levels, leading to aberrant keratinocyte proliferation and differentiation. Although certain clinical phenotypes, such as plaque psoriasis, are well defined, it is currently unclear whether there are molecular subtypes that might impact on prognosis or treatment outcomes.
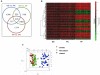
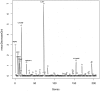
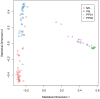
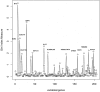
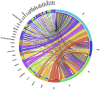
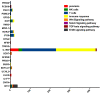
Results: We present a pipeline for patient stratification through a comprehensive analysis of gene expression in paired lesional and non-lesional psoriatic tissue samples, compared with controls, to establish differences in RNA expression patterns across all tissue types. Ensembles of decision tree predictors were employed to cluster psoriatic samples on the basis of gene expression patterns and reveal gene expression signatures that best discriminate molecular disease subtypes. This multi-stage procedure was applied to several published psoriasis studies and a comparison of gene expression patterns across datasets was performed.
Conclusion: Overall, classification of psoriasis gene expression patterns revealed distinct molecular sub-groups within the clinical phenotype of plaque psoriasis. Enrichment for TGFb and ErbB signaling pathways, noted in one of the two psoriasis subgroups, suggested that this group may be more amenable to therapies targeting these pathways. Our study highlights the potential biological relevance of using ensemble decision tree predictors to determine molecular disease subtypes, in what may initially appear to be a homogenous clinical group. The R code used in this paper is available upon request.
Figures
References
-
- Nestle FO, Kaplan DH, Barker J. Psoriasis. N Engl J Med. 2009;361(5):496–509. - PubMed
-
- Lebwohl M. Psoriasis. Lancet. 2003;361(9364):1197–1204. - PubMed
-
- Lowes MA, Bowcock AM, Krueger JG. Pathogenesis and therapy of psoriasis. Nature. 2007;445(7130):866–873. - PubMed
-
- Capon F, Di Meglio P, Szaub J, Prescott NJ, Dunster C, Baumber L, Timms K, Gutin A, Abkevic V, Burden AD. et al. Sequence variants in the genes for the interleukin-23 receptor (IL23R) and its ligand (IL12B) confer protection against psoriasis. Hum Genet. 2007;122(2):201–206. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

