Coupling methanol denaturation, immobilized trypsin digestion, and accurate mass and time tagging for liquid-chromatography-based shotgun proteomics of low nanogram amounts of RAW 264.7 cell lysate
- PMID: 22971241
- PMCID: PMC3477608
- DOI: 10.1021/ac3019608
Coupling methanol denaturation, immobilized trypsin digestion, and accurate mass and time tagging for liquid-chromatography-based shotgun proteomics of low nanogram amounts of RAW 264.7 cell lysate
Abstract
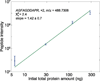
We report the shotgun proteomic analysis of mammalian cell lysates that contain low nanogram amounts of protein. Proteins were denatured using methanol, digested using immobilized trypsin, and analyzed by UPLC-ESI-MS/MS. The approach generated more peptides and higher sequence coverage for a mixture of three standard proteins than the use of free trypsin solution digestion of heat- or urea-denatured proteins. We prepared triplicate RAW 264.7 cell lysates that contained 6, 30, 120, and 300 ng of protein. An average of 2 ± 1, 23 ± 2, 134 ± 11, and 218 ± 26 proteins were detected for each sample size, respectively. The numbers of both protein and peptide IDs scaled linearly with the amount of sample taken for analysis. Our approach also outperformed traditional methods (free trypsin digestion of heat- or urea-denatured proteins) for 6-300 ng RAW 264.7 cell protein analysis in terms of number of peptides and proteins identified. The use of accurate mass and time (AMT) tags resulted in the identification of an additional 16 proteins based on 20 peptides from the 6 ng cell lysate prepared with our approach. When AMT analysis was performed for the 6 ng cell lysate prepared with traditional methods, no reasonable peptide signal could be obtained. In all cases, roughly ∼30% of the digested sample was taken for analysis, corresponding to the analysis of a 2 ng aliquot of homogenate from the 6 ng cell lysate.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

