Identification of chemotaxis sensory proteins for amino acids in Pseudomonas fluorescens Pf0-1 and their involvement in chemotaxis to tomato root exudate and root colonization
- PMID: 22972385
- PMCID: PMC4103555
- DOI: 10.1264/jsme2.me12005
Identification of chemotaxis sensory proteins for amino acids in Pseudomonas fluorescens Pf0-1 and their involvement in chemotaxis to tomato root exudate and root colonization
Abstract
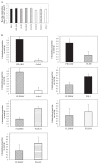
Pseudomonas fluorescens Pf0-1 showed positive chemotactic responses toward 20 commonly-occurring l-amino acids. Genomic analysis revealed that P. fluorescens Pf0-1 possesses three genes (Pfl01_0124, Pfl01_0354, and Pfl01_4431) homologous to the Pseudomonas aeruginosa PAO1 pctA gene, which has been identified as a chemotaxis sensory protein for amino acids. When Pf01_4431, Pfl01_0124, and Pfl01_0354 were introduced into the pctA pctB pctC triple mutant of P. aeruginosa PAO1, a mutant defective in chemotaxis to amino acids, its transformants showed chemotactic responses to 18, 16, and one amino acid, respectively. This result suggests that Pf01_4431, Pfl01_0124, and Pfl01_0354 are chemotaxis sensory proteins for amino acids and their genes were designated ctaA, ctaB, and ctaC, respectively. The ctaA ctaB ctaC triple mutant of P. fluorescens Pf0-1 showed only weak responses to Cys and Pro but no responses to the other 18 amino acids, indicating that CtaA, CtaB, and CtaC are major chemotaxis sensory proteins in P. fluorescens Pf0-1. Tomato root colonization by P. fluorescens strains was analyzed by gnotobiotic competitive root colonization assay. It was found that ctaA ctaB ctaC mutant was less competitive than the wild-type strain, suggesting that chemotaxis to amino acids, major components of root exudate, has an important role in root colonization by P. fluorescens Pf0-1. The ctaA ctaB ctaC triple mutant was more competitive than the cheA mutant of P. fluorescens Pf0-1, which is non-chemotactic, but motile. This result suggests that chemoattractants other than amino acids are also involved in root colonization by P. fluorescens Pf0-1.
Figures


References
-
- Barahona E, Navazo A, Martínez-Granero F, Zea-Bonilla T, Pérez-Jiménez RM, Martín M, Rivilla R. Pseudomonas fluorescens F113 mutant with enhanced competitive colonization ability and improved biocontrol activity against fungal root pathogens. Appl Environ Microbiol. 2011;77:5412–5419. - PMC - PubMed
-
- Bolwerk A, Lagopodi AL, Wijfjes AH, Lamers GE, Chin-A-Woeng TF, Lugtenberg BJ, Bloemberg GV. Interactions in the tomato rhizosphere of two Pseudomonas biocontrol strains with the phytopathogenic fungus Fusarium oxysporum f. sp. radicislycopersici. Mol Plant Microbe Interact. 2003;16:983–993. - PubMed

