SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells
- PMID: 22972397
- PMCID: PMC3828732
- DOI: 10.1038/nm.2964
SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells
Abstract
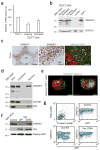
Unlike activated CD4(+) T cells, resting CD4(+) T cells are highly resistant to productive HIV-1 infection. Early after HIV-1 entry, a major block limits reverse transcription of incoming viral genomes. Here we show that the deoxynucleoside triphosphate triphosphohydrolase SAMHD1 prevents reverse transcription of HIV-1 RNA in resting CD4(+) T cells. SAMHD1 is abundantly expressed in resting CD4(+) T cells circulating in peripheral blood and residing in lymphoid organs. The early restriction to infection in unstimulated CD4(+) T cells is overcome by HIV-1 or HIV-2 virions into which viral Vpx is artificially or naturally packaged, respectively, or by addition of exogenous deoxynucleosides. Vpx-mediated proteasomal degradation of SAMHD1 and elevation of intracellular deoxynucleotide pools precede successful infection by Vpx-carrying HIV. Resting CD4(+) T cells from healthy donors following SAMHD1 silencing or from a patient with Aicardi-Goutières syndrome homozygous for a nonsense mutation in SAMHD1 were permissive for HIV-1 infection. Thus, SAMHD1 imposes an effective restriction to HIV-1 infection in the large pool of noncycling CD4(+) T cells in vivo. Bypassing SAMHD1 was insufficient for the release of viral progeny, implicating other barriers at later stages of HIV replication. Together, these findings may unveil new ways to interfere with the immune evasion and T cell immunopathology of pandemic HIV-1.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

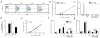

Comment in
-
SAMHD1 does it again, now in resting T cells.Nat Med. 2012 Nov;18(11):1611-2. doi: 10.1038/nm.2980. Nat Med. 2012. PMID: 23135508 Free PMC article.
References
-
- Ganesh L, et al. The gene product Murr1 restricts HIV-1 replication in resting CD4+ lymphocytes. Nature. 2003;426:853–857. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

