Transcriptional signatures as a disease-specific and predictive inflammatory biomarker for type 1 diabetes
- PMID: 22972474
- PMCID: PMC4265236
- DOI: 10.1038/gene.2012.41
Transcriptional signatures as a disease-specific and predictive inflammatory biomarker for type 1 diabetes
Abstract
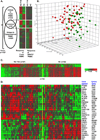
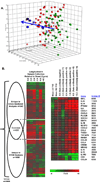
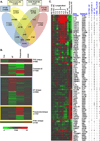
The complex milieu of inflammatory mediators associated with many diseases is often too dilute to directly measure in the periphery, necessitating development of more sensitive measurements suitable for mechanistic studies, earlier diagnosis, guiding therapeutic decisions and monitoring interventions. We previously demonstrated that plasma samples from recent-onset type 1 diabetes (RO T1D) patients induce a proinflammatory transcriptional signature in freshly drawn peripheral blood mononuclear cells (PBMCs) relative to that of unrelated healthy controls (HC). Here, using cryopreserved PBMC, we analyzed larger RO T1D and HC cohorts, examined T1D progression in pre-onset samples, and compared the RO T1D signature to those associated with three disorders characterized by airway infection and inflammation. The RO T1D signature, consisting of interleukin-1 cytokine family members, chemokines involved in immunocyte chemotaxis, immune receptors and signaling molecules, was detected during early pre-diabetes and found to resolve post-onset. The signatures associated with cystic fibrosis patients chronically infected with Pseudomonas aeruginosa, patients with confirmed bacterial pneumonia, and subjects with H1N1 influenza all reflected immunological activation, yet each were distinct from one another and negatively correlated with that of T1D. This study highlights the remarkable capacity of cells to serve as biosensors capable of sensitively and comprehensively differentiating immunological states.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Jenner RG, Young RA. Insights into host responses against pathogens from transcriptional profiling. Nat Rev Microbiol. 2005;3(4):281–294. - PubMed
-
- Medzhitov R, Janeway C., Jr Innate immune recognition: mechanisms and pathways. Immunol Rev. 2000;173:89–97. - PubMed
-
- Elson G, Dunn-Siegrist I, Daubeuf B, Pugin J. Contribution of Toll-like receptors to the innate immune response to Gram-negative and Gram-positive bacteria. Blood. 2007;109(4):1574–1583. - PubMed
-
- Underhill DM, Ozinsky A. Toll-like receptors: key mediators of microbe detection. Curr Opin Immunol. 2002;14(1):103–110. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

