Crystal structure of a preacylation complex of the β-lactamase inhibitor sulbactam bound to a sulfenamide bond-containing thiol-β-lactamase
- PMID: 22974281
- PMCID: PMC4037319
- DOI: 10.1021/ja3073676
Crystal structure of a preacylation complex of the β-lactamase inhibitor sulbactam bound to a sulfenamide bond-containing thiol-β-lactamase
Abstract
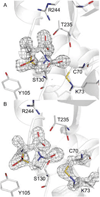
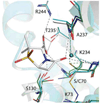
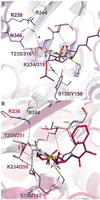
The rise of inhibitor-resistant and other β-lactamase variants is generating an interest in developing new β-lactamase inhibitors to complement currently available antibiotics. To gain insight into the chemistry of inhibitor recognition, we determined the crystal structure of the inhibitor preacylation complex of sulbactam, a clinical β-lactamase inhibitor, bound in the active site of the S70C variant of SHV-1 β-lactamase, a resistance enzyme that is normally present in Klebsiella pneumoniae. The S70C mutation was designed to affect the reactivity of that catalytic residue to allow for capture of the preacylation complex. Unexpectedly, the 1.45 Å resolution inhibitor complex structure revealed that residue C70 is involved in a sulfenamide bond with K73. Such a covalent bond is not present in the wild-type SHV-1 or in an apo S70C structure also determined in this study. This bond likely contributed significantly to obtaining the preacylation complex with sulbactam due to further decreased reactivity toward substrates. The intact sulbactam is positioned in the active site such that its carboxyl moiety interacts with R244, S130, and T235 and its carbonyl moiety is situated in the oxyanion hole. To our knowledge, in addition to being the first preacylation inhibitor β-lactamase complex, this is also the first observation of a sulfenamide bond between a cysteine and lysine in an active site. Not only could our results aid, therefore, structure-based inhibitor design efforts in class A β-lactamases, but the sulfenamide-bond forming approach to yield preacylation complexes could also be applied to other classes of β-lactamases and penicillin-binding proteins with the SXXK motif.
Figures

Similar articles
-
New Conformations of Acylation Adducts of Inhibitors of β-Lactamase from Mycobacterium tuberculosis.Biochemistry. 2019 Feb 19;58(7):997-1009. doi: 10.1021/acs.biochem.8b01085. Epub 2019 Jan 30. Biochemistry. 2019. PMID: 30632739 Free PMC article.
-
Crystal structures of KPC-2 β-lactamase in complex with 3-nitrophenyl boronic acid and the penam sulfone PSR-3-226.Antimicrob Agents Chemother. 2012 May;56(5):2713-8. doi: 10.1128/AAC.06099-11. Epub 2012 Feb 13. Antimicrob Agents Chemother. 2012. PMID: 22330909 Free PMC article.
-
Inhibition of Klebsiella β-Lactamases (SHV-1 and KPC-2) by Avibactam: A Structural Study.PLoS One. 2015 Sep 4;10(9):e0136813. doi: 10.1371/journal.pone.0136813. eCollection 2015. PLoS One. 2015. PMID: 26340563 Free PMC article.
-
SHV-type beta-lactamases.Curr Pharm Des. 1999 Nov;5(11):847-64. Curr Pharm Des. 1999. PMID: 10539992 Review.
-
Sulbactam/Durlobactam: First Approval.Drugs. 2023 Sep;83(13):1245-1252. doi: 10.1007/s40265-023-01920-6. Drugs. 2023. PMID: 37523122 Review.
Cited by
-
Avibactam and inhibitor-resistant SHV β-lactamases.Antimicrob Agents Chemother. 2015 Jul;59(7):3700-9. doi: 10.1128/AAC.04405-14. Epub 2015 Feb 17. Antimicrob Agents Chemother. 2015. PMID: 25691639 Free PMC article.
-
New Conformations of Acylation Adducts of Inhibitors of β-Lactamase from Mycobacterium tuberculosis.Biochemistry. 2019 Feb 19;58(7):997-1009. doi: 10.1021/acs.biochem.8b01085. Epub 2019 Jan 30. Biochemistry. 2019. PMID: 30632739 Free PMC article.
-
Redox Signaling by Reactive Electrophiles and Oxidants.Chem Rev. 2018 Sep 26;118(18):8798-8888. doi: 10.1021/acs.chemrev.7b00698. Epub 2018 Aug 27. Chem Rev. 2018. PMID: 30148624 Free PMC article. Review.
-
Heterogeneity in M. tuberculosis β-lactamase inhibition by Sulbactam.Nat Commun. 2023 Sep 7;14(1):5507. doi: 10.1038/s41467-023-41246-1. Nat Commun. 2023. PMID: 37679343 Free PMC article.
-
Widespread occurrence of covalent lysine-cysteine redox switches in proteins.Nat Chem Biol. 2022 Apr;18(4):368-375. doi: 10.1038/s41589-021-00966-5. Epub 2022 Feb 14. Nat Chem Biol. 2022. PMID: 35165445 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

