Functional insights from glutamate receptor ion channel structures
- PMID: 22974439
- PMCID: PMC4130219
- DOI: 10.1146/annurev-physiol-030212-183711
Functional insights from glutamate receptor ion channel structures
Abstract
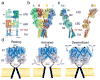
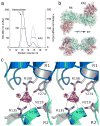
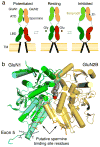
X-ray crystal structures for the soluble amino-terminal and ligand-binding domains of glutamate receptor ion channels, combined with a 3.6-Å-resolution structure of the full-length AMPA receptor GluA2 homotetramer, provide unique insights into the mechanisms of the assembly and function of glutamate receptor ion channels. Increasingly sophisticated biochemical, computational, and electrophysiological experiments are beginning to reveal the mechanism of action of partial agonists and suggest new models for the mechanism of action of allosteric modulators. Newly identified NMDA receptor ligands acting at novel sites offer hope for the development of subtype-selective modulators. The many unresolved issues include the role of the amino-terminal domain in AMPA receptor signaling and the mechanisms by which auxiliary proteins regulate receptor activity. The structural basis for ion permeation and ion channel block also remain areas of uncertainty, and despite substantial progress, molecular dynamics simulations have yet to reveal how glutamate binding opens the ion channel pore.
Figures

References
LITERATURE CITED
-
- Petralia RS, Wang YX, Wenthold RJ. Histological and ultrastructural localization of the kainate receptor subunits, KA2 and GluR6/7, in the rat nervous system using selective antipeptide antibodies. J Comp Neurol. 1994;349:85–110. - PubMed
Related articles
-
- Yuzaki M. Cbln1 and its family proteins in synapse formation and maintenance. Curr Opin Neurobiol. 2011;21:215–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

