Structure-guided engineering of Lactococcus lactis alcohol dehydrogenase LlAdhA for improved conversion of isobutyraldehyde to isobutanol
- PMID: 22974724
- PMCID: PMC3542407
- DOI: 10.1016/j.jbiotec.2012.08.008
Structure-guided engineering of Lactococcus lactis alcohol dehydrogenase LlAdhA for improved conversion of isobutyraldehyde to isobutanol
Abstract
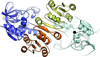
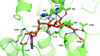
We have determined the X-ray crystal structures of the NADH-dependent alcohol dehydrogenase LlAdhA from Lactococcus lactis and its laboratory-evolved variant LlAdhA(RE1) at 1.9Å and 2.5Å resolution, respectively. LlAdhA(RE1), which contains three amino acid mutations (Y50F, I212T, and L264V), was engineered to increase the microbial production of isobutanol (2-methylpropan-1-ol) from isobutyraldehyde (2-methylpropanal). Structural comparison of LlAdhA and LlAdhA(RE1) indicates that the enhanced activity on isobutyraldehyde stems from increases in the protein's active site size, hydrophobicity, and substrate access. Further structure-guided mutagenesis generated a quadruple mutant (Y50F/N110S/I212T/L264V), whose KM for isobutyraldehyde is ∼17-fold lower and catalytic efficiency (kcat/KM) is ∼160-fold higher than wild-type LlAdhA. Combining detailed structural information and directed evolution, we have achieved significant improvements in non-native alcohol dehydrogenase activity that will facilitate the production of next-generation fuels such as isobutanol from renewable resources.
Copyright © 2012 Elsevier B.V. All rights reserved.
Conflict of interest statement
F.H.A. and P.M. are co-founders and shareholders of Gevo, Inc.
Figures

Similar articles
-
Corynebacterium glutamicum tailored for efficient isobutanol production.Appl Environ Microbiol. 2011 May;77(10):3300-10. doi: 10.1128/AEM.02972-10. Epub 2011 Mar 25. Appl Environ Microbiol. 2011. PMID: 21441331 Free PMC article.
-
Production of C4 and C5 branched-chain alcohols by engineered Escherichia. coli.J Ind Microbiol Biotechnol. 2015 Nov;42(11):1473-9. doi: 10.1007/s10295-015-1656-z. Epub 2015 Sep 8. J Ind Microbiol Biotechnol. 2015. PMID: 26350079
-
Only one of the two annotated Lactococcus lactis fabG genes encodes a functional beta-ketoacyl-acyl carrier protein reductase.Biochemistry. 2004 Sep 21;43(37):11782-9. doi: 10.1021/bi0487600. Biochemistry. 2004. PMID: 15362862
-
Engineering the isobutanol biosynthetic pathway in Escherichia coli by comparison of three aldehyde reductase/alcohol dehydrogenase genes.Appl Microbiol Biotechnol. 2010 Jan;85(3):651-7. doi: 10.1007/s00253-009-2085-6. Epub 2009 Jul 16. Appl Microbiol Biotechnol. 2010. PMID: 19609521 Free PMC article.
-
Crystallographic analysis and structure-guided engineering of NADPH-dependent Ralstonia sp. alcohol dehydrogenase toward NADH cosubstrate specificity.Biotechnol Bioeng. 2013 Nov;110(11):2803-14. doi: 10.1002/bit.24956. Epub 2013 Jul 1. Biotechnol Bioeng. 2013. PMID: 23686719
Cited by
-
Engineering new-to-nature biochemical conversions by combining fermentative metabolism with respiratory modules.Nat Commun. 2024 Aug 7;15(1):6725. doi: 10.1038/s41467-024-51029-x. Nat Commun. 2024. PMID: 39112480 Free PMC article.
-
Isobutanol production by combined in vivo and in vitro metabolic engineering.Metab Eng Commun. 2022 Oct 23;15:e00210. doi: 10.1016/j.mec.2022.e00210. eCollection 2022 Dec. Metab Eng Commun. 2022. PMID: 36325486 Free PMC article.
-
Comparative functional genomics identifies an iron-limited bottleneck in a Saccharomyces cerevisiae strain with a cytosolic-localized isobutanol pathway.Synth Syst Biotechnol. 2022 Mar 18;7(2):738-749. doi: 10.1016/j.synbio.2022.02.007. eCollection 2022 Jun. Synth Syst Biotechnol. 2022. PMID: 35387233 Free PMC article.
-
Substrate expansion of Geotrichum candidum alcohol dehydrogenase towards diaryl ketones by mutation.Appl Microbiol Biotechnol. 2024 Dec 27;108(1):545. doi: 10.1007/s00253-024-13375-0. Appl Microbiol Biotechnol. 2024. PMID: 39729095 Free PMC article.
-
Engineering the carbon and redox metabolism of Paenibacillus polymyxa for efficient isobutanol production.Microb Biotechnol. 2024 Mar;17(3):e14438. doi: 10.1111/1751-7915.14438. Microb Biotechnol. 2024. PMID: 38529712 Free PMC article.
References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:213–221. - PMC - PubMed
-
- Agarwal PK, Webb SP, Hammes-Schiffer S. Computational studies of the mechanism for proton and hydride transfer in liver alcohol dehydrogenase. J. Am. Chem. Soc. 2000;122:4803–4812. - PubMed
-
- Atsumi S, Hanai T, Liao JC. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451:86–89. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

