Psk1, an AGC kinase family member in fission yeast, is directly phosphorylated and controlled by TORC1 and functions as S6 kinase
- PMID: 22976295
- PMCID: PMC3575713
- DOI: 10.1242/jcs.111146
Psk1, an AGC kinase family member in fission yeast, is directly phosphorylated and controlled by TORC1 and functions as S6 kinase
Abstract
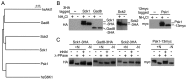
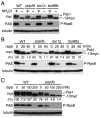
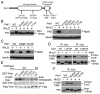
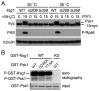
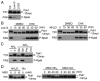
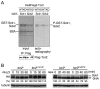
Target of rapamycin (TOR), an evolutionarily conserved serine/threonine protein kinase, plays pivotal roles in several important cellular processes in eukaryotes. In the fission yeast Schizosaccharomyces pombe, TOR complex 1 (TORC1), which includes Tor2 as a catalytic subunit, manages the switch between cell proliferation and differentiation by sensing nutrient availability. However, little is known about the direct target of TORC1 that plays key roles in nutrient-dependent TORC1 signaling in fission yeast. Here we report that in fission yeast, three AGC kinase family members, named Psk1, Sck1 and Sck2, which exhibit high homology with human S6K1, are phosphorylated under nutrient-rich conditions and are dephosphorylated by starvation conditions. Among these, Psk1 is necessary for phosphorylation of ribosomal protein S6. Furthermore, Psk1 phosphorylation is regulated by TORC1 in nutrient-dependent and rapamycin-sensitive manners in vivo. Three conserved regulatory motifs (the activation loop, the hydrophobic and the turn motifs) in Psk1 are phosphorylated and these modifications are required for Psk1 activity. In particular, phosphorylation of the hydrophobic motif is catalyzed by TORC1 in vivo and in vitro. Ksg1, a homolog of PDK1, is also important for Psk1 phosphorylation in the activation loop and for its activity. The TORC1 components Pop3, Toc1 and Tco89, are dispensable for Psk1 regulation, but disruption of pop3(+) causes an increase in the sensitivity of TORC1 to rapamycin. Taken together, these results provide convincing evidence that TORC1/Psk1/Rps6 constitutes a nutrient-dependent signaling pathway in fission yeast.
Figures


Similar articles
-
Fission yeast TORC1 regulates phosphorylation of ribosomal S6 proteins in response to nutrients and its activity is inhibited by rapamycin.J Cell Sci. 2010 Mar 1;123(Pt 5):777-86. doi: 10.1242/jcs.060319. Epub 2010 Feb 9. J Cell Sci. 2010. PMID: 20144990 Free PMC article.
-
Fission yeast TOR complex 1 phosphorylates Psk1 through an evolutionarily conserved interaction mediated by the TOS motif.J Cell Sci. 2021 Oct 1;134(19):jcs258865. doi: 10.1242/jcs.258865. Epub 2021 Oct 12. J Cell Sci. 2021. PMID: 34499159 Free PMC article.
-
The cytosolic form of aspartate aminotransferase is required for full activation of TOR complex 1 in fission yeast.J Biol Chem. 2019 Nov 29;294(48):18244-18255. doi: 10.1074/jbc.RA119.010101. Epub 2019 Oct 22. J Biol Chem. 2019. PMID: 31641022 Free PMC article.
-
TORC1-Dependent Phosphorylation Targets in Fission Yeast.Biomolecules. 2017 Jul 3;7(3):50. doi: 10.3390/biom7030050. Biomolecules. 2017. PMID: 28671615 Free PMC article. Review.
-
Fission Yeast TORC2 Signaling Pathway Ensures Cell Proliferation under Glucose-Limited, Nitrogen-Replete Conditions.Biomolecules. 2021 Oct 6;11(10):1465. doi: 10.3390/biom11101465. Biomolecules. 2021. PMID: 34680098 Free PMC article. Review.
Cited by
-
Multiple crosstalk between TOR and the cell integrity MAPK signaling pathway in fission yeast.Sci Rep. 2016 Nov 23;6:37515. doi: 10.1038/srep37515. Sci Rep. 2016. PMID: 27876895 Free PMC article.
-
Distinct functional relevance of dynamic GTPase cysteine methylation in fission yeast.Sci Rep. 2017 Jul 20;7(1):6057. doi: 10.1038/s41598-017-06053-x. Sci Rep. 2017. PMID: 28729673 Free PMC article.
-
Caffeine Stabilises Fission Yeast Wee1 in a Rad24-Dependent Manner but Attenuates Its Expression in Response to DNA Damage.Microorganisms. 2020 Sep 30;8(10):1512. doi: 10.3390/microorganisms8101512. Microorganisms. 2020. PMID: 33008060 Free PMC article.
-
The Greatwall-Endosulfine Switch Accelerates Autophagic Flux during the Cell Divisions Leading to G1 Arrest and Entry into Quiescence in Fission Yeast.Int J Mol Sci. 2022 Dec 21;24(1):148. doi: 10.3390/ijms24010148. Int J Mol Sci. 2022. PMID: 36613592 Free PMC article.
-
The Central FacilitaTOR: Coordinating Transcription and Translation in Eukaryotes.Int J Mol Sci. 2025 Mar 21;26(7):2845. doi: 10.3390/ijms26072845. Int J Mol Sci. 2025. PMID: 40243440 Free PMC article. Review.
References
-
- Bähler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A., 3rd, Steever A. B., Wach A., Philippsen P., Pringle J. R. (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

