The SUMO ligase PIAS1 regulates UV-induced apoptosis by recruiting Daxx to SUMOylated foci
- PMID: 22976298
- PMCID: PMC3575712
- DOI: 10.1242/jcs.110825
The SUMO ligase PIAS1 regulates UV-induced apoptosis by recruiting Daxx to SUMOylated foci
Abstract
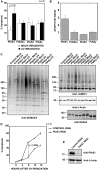
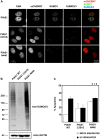
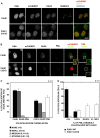
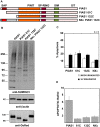
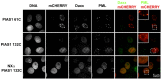
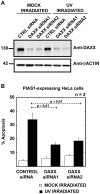
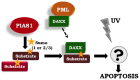
The small ubiquitin-like modifier (SUMO) ligase PIAS1 (Protein Inhibitor of Activated Stat-1) has been shown to play a role in cellular stress response by SUMOylating several proteins that are involved in DNA repair, apoptosis and transcription. In this paper, we show that PIAS1 regulates ultraviolet (UV)-induced apoptosis by recruiting Death-associated protein 6 (Daxx) to PIAS1-generated SUMO-foci. Cells that ectopically express PIAS1, but not other PIASes, show increased sensitivity to UV irradiation, suggesting that PIAS1 has a distinct function in UV-dependent apoptosis. Domain analysis of PIAS1 indicates that both PIAS1 SUMO-ligase activity and the specific localization of PIAS1 through its N-terminal and C-terminal domains are essential for UV-induced cell death. Daxx colocalizes with PIAS1-generated SUMOylated foci, and the reduction of Daxx using RNAi alleviates UV-induced apoptosis in PIAS1-expressing cells. PIAS1-mediated recruitment of Daxx and apoptosis following UV irradiation are dependent upon the Daxx C-terminal SUMO-interacting motif (SIM). Overall, our data suggest that the pro-apoptotic protein Daxx specifically interacts with one or more substrates SUMOylated by PIAS1 and this interaction leads to apoptosis following UV irradiation.
Figures

Similar articles
-
Characterizing the N- and C-terminal Small ubiquitin-like modifier (SUMO)-interacting motifs of the scaffold protein DAXX.J Biol Chem. 2011 Jun 3;286(22):19816-29. doi: 10.1074/jbc.M111.231647. Epub 2011 Mar 7. J Biol Chem. 2011. PMID: 21383010 Free PMC article.
-
SUMO Ligase Protein Inhibitor of Activated STAT1 (PIAS1) Is a Constituent Promyelocytic Leukemia Nuclear Body Protein That Contributes to the Intrinsic Antiviral Immune Response to Herpes Simplex Virus 1.J Virol. 2016 Jun 10;90(13):5939-5952. doi: 10.1128/JVI.00426-16. Print 2016 Jul 1. J Virol. 2016. PMID: 27099310 Free PMC article.
-
Structural and functional roles of Daxx SIM phosphorylation in SUMO paralog-selective binding and apoptosis modulation.Mol Cell. 2011 Apr 8;42(1):62-74. doi: 10.1016/j.molcel.2011.02.022. Mol Cell. 2011. PMID: 21474068
-
Daxx mediates SUMO-dependent transcriptional control and subnuclear compartmentalization.Biochem Soc Trans. 2007 Dec;35(Pt 6):1397-400. doi: 10.1042/BST0351397. Biochem Soc Trans. 2007. PMID: 18031230 Review.
-
The Dynamic Regulation of Daxx-Mediated Transcriptional Inhibition by SUMO and PML NBs.Int J Mol Sci. 2025 Jul 12;26(14):6703. doi: 10.3390/ijms26146703. Int J Mol Sci. 2025. PMID: 40724953 Free PMC article. Review.
Cited by
-
Necdin promotes ubiquitin-dependent degradation of PIAS1 SUMO E3 ligase.PLoS One. 2014 Jun 9;9(6):e99503. doi: 10.1371/journal.pone.0099503. eCollection 2014. PLoS One. 2014. PMID: 24911587 Free PMC article.
-
PIAS1-mediated SUMOylation of influenza A virus PB2 restricts viral replication and virulence.PLoS Pathog. 2022 Apr 4;18(4):e1010446. doi: 10.1371/journal.ppat.1010446. eCollection 2022 Apr. PLoS Pathog. 2022. PMID: 35377920 Free PMC article.
-
SUMO and Parkinson's disease.Neuromolecular Med. 2013 Dec;15(4):737-59. doi: 10.1007/s12017-013-8259-5. Epub 2013 Aug 25. Neuromolecular Med. 2013. PMID: 23979994 Review.
-
Truncated PARP1 mediates ADP-ribosylation of RNA polymerase III for apoptosis.Cell Discov. 2022 Jan 18;8(1):3. doi: 10.1038/s41421-021-00355-1. Cell Discov. 2022. PMID: 35039483 Free PMC article.
-
Chromosomal localization of Ewing sarcoma EWSR1/FLI1 protein promotes the induction of aneuploidy.J Biol Chem. 2021 Jan-Jun;296:100164. doi: 10.1074/jbc.RA120.014328. Epub 2020 Dec 10. J Biol Chem. 2021. PMID: 33293370 Free PMC article.
References
-
- Carpenter A. E., Jones T. R., Lamprecht M. R., Clarke C., Kang I. H., Friman O., Guertin D. A., Chang J. H., Lindquist R. A., Moffat J.et al. (2006). CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 10.1186/gb-2006-7-10-r100 - DOI - PMC - PubMed
-
- Chang C. C., Naik M. T., Huang Y. S., Jeng J. C., Liao P. H., Kuo H. Y., Ho C. C., Hsieh Y. L., Lin C. H., Huang N. J.et al. (2011). Structural and functional roles of Daxx SIM phosphorylation in SUMO paralog-selective binding and apoptosis modulation. Mol. Cell 42, 62–74 10.1016/j.molcel.2011.02.022 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

