Dual fluorescence detection of protein and RNA in Drosophila tissues
- PMID: 22976352
- PMCID: PMC4821427
- DOI: 10.1038/nprot.2012.105
Dual fluorescence detection of protein and RNA in Drosophila tissues
Abstract
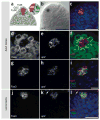
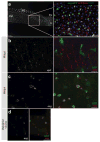
Detection of RNAs by in situ hybridization (ISH) is a well-established technique that permits the study of specific RNA expression patterns in tissues; however, not all tissues are equally amenable to staining using the same procedure. Here we describe a protocol that combines whole-mount immunofluorescence (IF) and fluorescence in situ hybridization (FISH) for the simultaneous detection of specific RNA transcripts and proteins, greatly enhancing the spatial resolution of RNA expression in complex, intact fly tissues. To date, we have successfully used this protocol in adult testis, larval male gonads, adult intestine and Malpighian tubules. IF is conducted in RNase-free solutions, prior to the harsh conditions of FISH, in order to preserve protein antigenicity within dissected tissues. Separate protocols are described for mRNA and miRNA detection, which are based on robust digoxigenin (DIG) RNA and locked nucleic acid (LNA) probes, respectively. The combined IF-FISH procedure can be completed in 2 d for miRNA detection and 4 d for mRNA detection. Although optimized for Drosophila, this IF-FISH protocol should be adaptable to a wide variety of organisms, tissues, antibodies and probes, thus providing a reliable and simple means to compare RNA and protein abundance and localization.
Conflict of interest statement
Figures


References
-
- Speel EJ, Hopman AH, Komminoth P. Amplification methods to increase the sensitivity of in situ hybridization: play card(s) J Histochem Cytochem. 1999;47:281–288. - PubMed
-
- Speel EJ, Saremaslani P, Roth J, Hopman AH, Komminoth P. Improved mRNA in situ hybridization on formaldehyde-fixed and paraffin-embedded tissue using signal amplification with different haptenized tyramides. Histochem Cell Biol. 1998;110:571–577. - PubMed
-
- Baldino F, Chesselet MF, Lewis ME., Jr High-resolution in situ hybridization histochemistry. Methods Enzymol. 1989;168:761–777. - PubMed
-
- Springer JE, Robbins E, Gwag BJ, Lewis ME, Baldino F., Jr Non-radioactive detection of nerve growth factor receptor (NGFR) mRNA in rat brain using in situ hybridization histochemistry. J Histochem Cytochem. 1991;39:231–234. - PubMed
-
- Bobrow MN, Litt GJ, Shaughnessy KJ, Mayer PC, Conlon J. The use of catalyzed reporter deposition as a means of signal amplification in a variety of formats. J Immunol Methods. 1992;150:145–149. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

