Trim17-mediated ubiquitination and degradation of Mcl-1 initiate apoptosis in neurons
- PMID: 22976837
- PMCID: PMC3554334
- DOI: 10.1038/cdd.2012.124
Trim17-mediated ubiquitination and degradation of Mcl-1 initiate apoptosis in neurons
Abstract
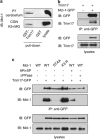
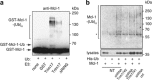
Short-term proteasome inhibition has been shown to prevent neuronal apoptosis. However, the key pro-survival proteins that must be degraded for triggering neuronal death are mostly unknown. Here, we show that Mcl-1, an anti-apoptotic Bcl-2 family member, is degraded by the proteasome during neuronal apoptosis. Using primary cultures of cerebellar granule neurons deprived of serum and KCl, we found that ubiquitination and proteasomal degradation of Mcl-1 depended on its prior phosphorylation by GSK3, providing the first insight into post-translational regulation of Mcl-1 in neurons. In a previous study, we have reported that the E3 ubiquitin-ligase Trim17 is both necessary and sufficient for neuronal apoptosis. Here, we identified Trim17 as a novel E3 ubiquitin-ligase for Mcl-1. Indeed, Trim17 co-immunoprecipitated with Mcl-1. Trim17 ubiquitinated Mcl-1 in vitro. Overexpression of Trim17 decreased the protein level of Mcl-1 in a phosphorylation- and proteasome-dependent manner. Finally, knock down of Trim17 expression reduced both ubiquitination and degradation of Mcl-1 in neurons. Moreover, impairment of Mcl-1 phosphorylation, by kinase inhibition or point mutations, not only decreased ubiquitination and degradation of Mcl-1, but also blocked the physical interaction between Trim17 and Mcl-1. As this stabilization of Mcl-1 increased its neuroprotective effect, our data strongly suggest that Trim17-mediated ubiquitination and degradation of Mcl-1 is necessary for initiating neuronal death.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

