Genetic characteristics of drug-resistant Vibrio cholerae O1 causing endemic cholera in Dhaka, 2006-2011
- PMID: 22977073
- PMCID: PMC4080733
- DOI: 10.1099/jmm.0.049635-0
Genetic characteristics of drug-resistant Vibrio cholerae O1 causing endemic cholera in Dhaka, 2006-2011
Abstract
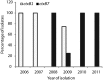
Vibrio cholerae O1 biotype El Tor (ET), causing the seventh cholera pandemic, was recently replaced in Bangladesh by an altered ET possessing ctxB of the Classical (CL) biotype, which caused the first six cholera pandemics. In the present study, V. cholerae O1 strains associated with endemic cholera in Dhaka between 2006 and 2011 were analysed for major phenotypic and genetic characteristics. Of 54 representative V. cholerae isolates tested, all were phenotypically ET and showed uniform resistance to trimethoprim/sulfamethoxazole (SXT) and furazolidone (FR). Resistance to tetracycline (TE) and erythromycin (E) showed temporal fluctuation, varying from year to year, while all isolates were susceptible to gentamicin (CN) and ciprofloxacin (CIP). Year-wise data revealed erythromycin resistance to be 33.3 % in 2006 and 11 % in 2011, while tetracycline resistance accounted for 33, 78, 0, 100 and 27 % in 2006, 2007, 2008, 2009 and 2010, respectively; interestingly, all isolates tested were sensitive to TE in 2011, as observed in 2008. All V. cholerae isolates tested possessed genetic elements such as SXT, ctxAB, tcpA(ET), rstR(ET) and rtxC; none had IntlI (Integron I). Double mismatch amplification mutation assay (DMAMA)-PCR followed by DNA sequencing and analysis of the ctxB gene revealed a point mutation at position 58 (C→A), which has resulted in an amino acid substitution from histidine (H) to asparagine (N) at position 20 (genotype 7) since 2008. Although the multi-resistant strains having tetracycline resistance showed minor genetic divergence, V. cholerae strains were clonal, as determined by a PFGE (NotI)-based dendrogram. This study shows 2008-2010 to be the time of transition from ctxB genotype 1 to genotype 7 in V. cholerae ET causing endemic cholera in Dhaka, Bangladesh.
Figures


References
-
- Alam M., Hasan N. A., Sadique A., Bhuiyan N. A., Ahmed K. U., Nusrin S., Nair G. B., Siddique A. K., Sack R. B. & other authors (2006). Seasonal cholera caused by Vibrio cholerae serogroups O1 and O139 in the coastal aquatic environment of Bangladesh. Appl Environ Microbiol 72, 4096–4104 10.1128/AEM.00066-06 - DOI - PMC - PubMed
-
- Alam M., Sultana M., Nair G. B., Siddique A. K., Hasan N. A., Sack R. B., Sack D. A., Ahmed K. U., Sadique A. & other authors (2007). Viable but nonculturable Vibrio cholerae O1 in biofilms in the aquatic environment and their role in cholera transmission. Proc Natl Acad Sci U S A 104, 17801–17806 10.1073/pnas.0705599104 - DOI - PMC - PubMed
-
- Alam M., Nusrin S., Islam A., Bhuiyan N. A., Rahim N., Delgado G., Morales R., Mendez J. L., Navarro A. & other authors (2010). Cholera between 1991 and 1997 in Mexico was associated with infection by classical, El Tor, and El Tor variants of Vibrio cholerae. J Clin Microbiol 48, 3666–3674 10.1128/JCM.00866-10 - DOI - PMC - PubMed
-
- Bauer A. W., Kirby W. M., Sherris J. C., Turck M. (1966). Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 45, 493–496 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

