Substitutions in the Escherichia coli RNA polymerase inhibitor T7 Gp2 that allow inhibition of transcription when the primary interaction interface between Gp2 and RNA polymerase becomes compromised
- PMID: 22977089
- PMCID: PMC3541766
- DOI: 10.1099/mic.0.062547-0
Substitutions in the Escherichia coli RNA polymerase inhibitor T7 Gp2 that allow inhibition of transcription when the primary interaction interface between Gp2 and RNA polymerase becomes compromised
Abstract
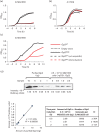
The Escherichia coli-infecting bacteriophage T7 encodes a 7 kDa protein, called Gp2, which is a potent inhibitor of the host RNA polymerase (RNAp). Gp2 is essential for T7 phage development. The interaction site for Gp2 on the E. coli RNAp is the β' jaw domain, which is part of the DNA binding channel. The binding of Gp2 to the β' jaw antagonizes several steps associated with interactions between the RNAp and promoter DNA, leading to inhibition of transcription at the open promoter complex formation step. In the structure of the complex formed between Gp2 and a fragment of the β' jaw, amino acid residues in the β3 strand of Gp2 contribute to the primary interaction interface with the β' jaw. The 7009 E. coli strain is resistant to T7 because it carries a charge reversal point mutation in the β' jaw that prevents Gp2 binding. However, a T7 phage encoding a mutant form of Gp2, called Gp2(β), which carries triple amino acid substitutions E24K, F27Y and R56C, can productively infect this strain. By studying the molecular basis of inhibition of RNAp from the 7009 strain by Gp2(β), we provide several lines of evidence that the E24K and F27Y substitutions facilitate an interaction with RNAp when the primary interaction interface with the β' jaw is compromised. The proposed additional interaction interface between RNAp and Gp2 may contribute to the multipronged mechanism of transcription inhibition by Gp2.
Figures





References
-
- Cámara B., Liu M., Reynolds J., Shadrin A., Liu B., Kwok K., Simpson P., Weinzierl R., Severinov K. & other authors (2010). T7 phage protein Gp2 inhibits the Escherichia coli RNA polymerase by antagonizing stable DNA strand separation near the transcription start site. Proc Natl Acad Sci U S A 107, 2247–2252. 10.1073/pnas.0907908107 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

