Evolution of conjugation and type IV secretion systems
- PMID: 22977114
- PMCID: PMC3548315
- DOI: 10.1093/molbev/mss221
Evolution of conjugation and type IV secretion systems
Abstract
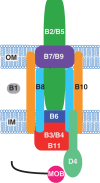
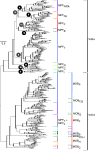
Genetic exchange by conjugation is responsible for the spread of resistance, virulence, and social traits among prokaryotes. Recent works unraveled the functioning of the underlying type IV secretion systems (T4SS) and its distribution and recruitment for other biological processes (exaptation), notably pathogenesis. We analyzed the phylogeny of key conjugation proteins to infer the evolutionary history of conjugation and T4SS. We show that single-stranded DNA (ssDNA) and double-stranded DNA (dsDNA) conjugation, while both based on a key AAA(+) ATPase, diverged before the last common ancestor of bacteria. The two key ATPases of ssDNA conjugation are monophyletic, having diverged at an early stage from dsDNA translocases. Our data suggest that ssDNA conjugation arose first in diderm bacteria, possibly Proteobacteria, and then spread to other bacterial phyla, including bacterial monoderms and Archaea. Identifiable T4SS fall within the eight monophyletic groups, determined by both taxonomy and structure of the cell envelope. Transfer to monoderms might have occurred only once, but followed diverse adaptive paths. Remarkably, some Firmicutes developed a new conjugation system based on an atypical relaxase and an ATPase derived from a dsDNA translocase. The observed evolutionary rates and patterns of presence/absence of specific T4SS proteins show that conjugation systems are often and independently exapted for other functions. This work brings a natural basis for the classification of all kinds of conjugative systems, thus tackling a problem that is growing as fast as genomic databases. Our analysis provides the first global picture of the evolution of conjugation and shows how a self-transferrable complex multiprotein system has adapted to different taxa and often been recruited by the host. As conjugation systems became specific to certain clades and cell envelopes, they may have biased the rate and direction of gene transfer by conjugation within prokaryotes.
Figures






Similar articles
-
In Situ Visualization of the pKM101-Encoded Type IV Secretion System Reveals a Highly Symmetric ATPase Energy Center.mBio. 2021 Oct 26;12(5):e0246521. doi: 10.1128/mBio.02465-21. Epub 2021 Oct 12. mBio. 2021. PMID: 34634937 Free PMC article.
-
Characterization of ConE, the VirB4 Homolog of the Integrative and Conjugative Element ICEBs1 of Bacillus subtilis.J Bacteriol. 2023 Jun 27;205(6):e0003323. doi: 10.1128/jb.00033-23. Epub 2023 May 23. J Bacteriol. 2023. PMID: 37219457 Free PMC article.
-
A bacterial toxin-antitoxin module is the origin of inter-bacterial and inter-kingdom effectors of Bartonella.PLoS Genet. 2017 Oct 26;13(10):e1007077. doi: 10.1371/journal.pgen.1007077. eCollection 2017 Oct. PLoS Genet. 2017. PMID: 29073136 Free PMC article.
-
Towards an integrated model of bacterial conjugation.FEMS Microbiol Rev. 2015 Jan;39(1):81-95. doi: 10.1111/1574-6976.12085. Epub 2014 Dec 4. FEMS Microbiol Rev. 2015. PMID: 25154632 Review.
-
The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification.Int J Syst Evol Microbiol. 2002 Jan;52(Pt 1):7-76. doi: 10.1099/00207713-52-1-7. Int J Syst Evol Microbiol. 2002. PMID: 11837318 Review.
Cited by
-
Origins of transfer establish networks of functional dependencies for plasmid transfer by conjugation.Nucleic Acids Res. 2023 Apr 24;51(7):3001-3016. doi: 10.1093/nar/gkac1079. Nucleic Acids Res. 2023. PMID: 36442505 Free PMC article.
-
Genetic and Functional Characterization of a Salicylate 1-monooxygenase Located on an Integrative and Conjugative Element (ICE) in Pseudomonas stutzeri AJR13.J Microbiol. 2023 Dec;61(12):1025-1032. doi: 10.1007/s12275-023-00093-x. Epub 2023 Dec 15. J Microbiol. 2023. PMID: 38100000
-
Functional amyloids promote retention of public goods in bacteria.Proc Biol Sci. 2019 May 29;286(1903):20190709. doi: 10.1098/rspb.2019.0709. Epub 2019 May 29. Proc Biol Sci. 2019. PMID: 31138071 Free PMC article.
-
Secretome of obligate intracellular Rickettsia.FEMS Microbiol Rev. 2015 Jan;39(1):47-80. doi: 10.1111/1574-6976.12084. Epub 2014 Dec 4. FEMS Microbiol Rev. 2015. PMID: 25168200 Free PMC article. Review.
-
Phylogeny of the Varidnaviria Morphogenesis Module: Congruence and Incongruence With the Tree of Life and Viral Taxonomy.Front Microbiol. 2021 Jul 16;12:704052. doi: 10.3389/fmicb.2021.704052. eCollection 2021. Front Microbiol. 2021. PMID: 34349745 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

