Circular retrotransposition products generated by a LINE retrotransposon
- PMID: 22977178
- PMCID: PMC3510499
- DOI: 10.1093/nar/gks859
Circular retrotransposition products generated by a LINE retrotransposon
Abstract
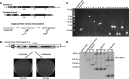
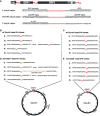
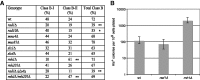
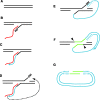
Non-long terminal repeat (non-LTR) retrotransposons are highly abundant elements that are present in chromosomes throughout the eukaryotic domain of life. The long interspersed nuclear element (LINE-1) (L1) clade of non-LTR retrotransposons has been particularly successful in mammals, accounting for 30-40% of human genome sequence. The current model of LINE retrotransposition, target-primed reverse transcription, culminates in a chromosomally integrated end product. Using a budding yeast model of non-LTR retrotransposition, we show that in addition to producing these 'classical', chromosomally integrated products, a fungal L1 clade member (Zorro3) can generate abundant, RNA-derived episomal products. Genetic evidence suggests that these products are likely to be formed via a variation of target-primed reverse transcription. These episomal products are a previously unseen alternative fate of LINE retrotransposition, and may represent an unexpected source for de novo retrotransposition.
Figures



Similar articles
-
A yeast model for target-primed (non-LTR) retrotransposition.BMC Genomics. 2007 Aug 3;8:263. doi: 10.1186/1471-2164-8-263. BMC Genomics. 2007. PMID: 17683538 Free PMC article.
-
LINE-like retrotransposition in Saccharomyces cerevisiae.Genetics. 2009 Jan;181(1):301-11. doi: 10.1534/genetics.108.096636. Epub 2008 Oct 28. Genetics. 2009. PMID: 18957700 Free PMC article.
-
Structural features and mechanism of translocation of non-LTR retrotransposons in Candida albicans.Virulence. 2014 Feb 15;5(2):245-52. doi: 10.4161/viru.27278. Epub 2013 Dec 6. Virulence. 2014. PMID: 24317340 Free PMC article. Review.
-
LINE Retrotransposition Assays in Saccharomyces cerevisiae.Methods Mol Biol. 2016;1400:131-7. doi: 10.1007/978-1-4939-3372-3_9. Methods Mol Biol. 2016. PMID: 26895051
-
Nucleic acid chaperone properties of ORF1p from the non-LTR retrotransposon, LINE-1.RNA Biol. 2010 Nov-Dec;7(6):706-11. doi: 10.4161/rna.7.6.13766. Epub 2010 Nov 1. RNA Biol. 2010. PMID: 21045547 Free PMC article. Review.
Cited by
-
LINEs of evidence: noncanonical DNA replication as an epigenetic determinant.Biol Direct. 2013 Sep 13;8:22. doi: 10.1186/1745-6150-8-22. Biol Direct. 2013. PMID: 24034780 Free PMC article. Review.
-
Potential Role of Accessory Domains in Polyproteins Encoded by Retrotransposons in Anti-viral Defense of Host Cells.Front Microbiol. 2019 Jan 4;9:3193. doi: 10.3389/fmicb.2018.03193. eCollection 2018. Front Microbiol. 2019. PMID: 30687243 Free PMC article. No abstract available.
-
Roles for retrotransposon insertions in human disease.Mob DNA. 2016 May 6;7:9. doi: 10.1186/s13100-016-0065-9. eCollection 2016. Mob DNA. 2016. PMID: 27158268 Free PMC article. Review.
-
Restricting retrotransposons: a review.Mob DNA. 2016 Aug 11;7:16. doi: 10.1186/s13100-016-0070-z. eCollection 2016. Mob DNA. 2016. PMID: 27525044 Free PMC article. Review.
-
Advances in Molecular Tools and In Vivo Models for the Study of Human Fungal Pathogenesis.Microorganisms. 2020 May 26;8(6):803. doi: 10.3390/microorganisms8060803. Microorganisms. 2020. PMID: 32466582 Free PMC article. Review.
References
-
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
-
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. - PubMed
-
- Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. - PubMed
-
- Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, Burch PE, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

