The Saccharomyces cerevisiae W303-K6001 cross-platform genome sequence: insights into ancestry and physiology of a laboratory mutt
- PMID: 22977733
- PMCID: PMC3438534
- DOI: 10.1098/rsob.120093
The Saccharomyces cerevisiae W303-K6001 cross-platform genome sequence: insights into ancestry and physiology of a laboratory mutt
Abstract
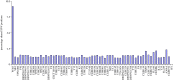
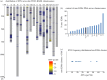
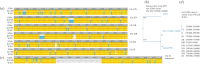
Saccharomyces cerevisiae strain W303 is a widely used model organism. However, little is known about its genetic origins, as it was created in the 1970s from crossing yeast strains of uncertain genealogy. To obtain insights into its ancestry and physiology, we sequenced the genome of its variant W303-K6001, a yeast model of ageing research. The combination of two next-generation sequencing (NGS) technologies (Illumina and Roche/454 sequencing) yielded an 11.8 Mb genome assembly at an N50 contig length of 262 kb. Although sequencing was substantially more precise and sensitive than whole-genome tiling arrays, both NGS platforms produced a number of false positives. At a 378× average coverage, only 74 per cent of called differences to the S288c reference genome were confirmed by both techniques. The consensus W303-K6001 genome differs in 8133 positions from S288c, predicting altered amino acid sequence in 799 proteins, including factors of ageing and stress resistance. The W303-K6001 (85.4%) genome is virtually identical (less than equal to 0.5 variations per kb) to S288c, and thus originates in the same ancestor. Non-S288c regions distribute unequally over the genome, with chromosome XVI the most (99.6%) and chromosome XI the least (54.5%) S288c-like. Several of these clusters are shared with Σ1278B, another widely used S288c-related model, indicating that these strains share a second ancestor. Thus, the W303-K6001 genome pictures details of complex genetic relationships between the model strains that date back to the early days of experimental yeast genetics. Moreover, this study underlines the necessity of combining multiple NGS and genome-assembling techniques for achieving accurate variant calling in genomic studies.
Keywords: mapping; next-generation sequencing; phylogeny reconstruction; yeast models.
Figures
References
-
- Breitenbach M, et al. 2012. The role of mitochondria in the aging processes of yeast. Subcell. Biochem. 57, 55–7810.1007/978-94-007-2561-4_3 (doi:10.1007/978-94-007-2561-4_3) - DOI - DOI - PubMed
-
- Partridge L. 2011. Some highlights of research on aging with invertebrates, 2010. Aging Cell 10, 5–910.1111/j.1474-9726.2010.00649.x (doi:10.1111/j.1474-9726.2010.00649.x) - DOI - DOI - PMC - PubMed
-
- Fabrizio P, Longo VD. 2003. The chronological life span of Saccharomyces cerevisiae. Aging Cell 2, 73–8110.1046/j.1474-9728.2003.00033.x (doi:10.1046/j.1474-9728.2003.00033.x) - DOI - DOI - PubMed
-
- Mortimer RK, Johnston JR. 1959. Life span of individual yeast cells. Nature 183, 1751–175210.1038/1831751a0 (doi:10.1038/1831751a0) - DOI - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
