Diversity and evolution of multiple orc/cdc6-adjacent replication origins in haloarchaea
- PMID: 22978470
- PMCID: PMC3528665
- DOI: 10.1186/1471-2164-13-478
Diversity and evolution of multiple orc/cdc6-adjacent replication origins in haloarchaea
Abstract
Background: While multiple replication origins have been observed in archaea, considerably less is known about their evolutionary processes. Here, we performed a comparative analysis of the predicted (proved in part) orc/cdc6-associated replication origins in 15 completely sequenced haloarchaeal genomes to investigate the diversity and evolution of replication origins in halophilic Archaea.
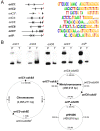
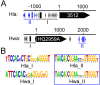
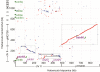
Results: Multiple orc/cdc6-associated replication origins were predicted in all of the analyzed haloarchaeal genomes following the identification of putative ORBs (origin recognition boxes) that are associated with orc/cdc6 genes. Five of these predicted replication origins in Haloarcula hispanica were experimentally confirmed via autonomous replication activities. Strikingly, several predicted replication origins in H. hispanica and Haloarcula marismortui are located in the distinct regions of their highly homologous chromosomes, suggesting that these replication origins might have been introduced as parts of new genomic content. A comparison of the origin-associated Orc/Cdc6 homologs and the corresponding predicted ORB elements revealed that the replication origins in a given haloarchaeon are quite diverse, while different haloarchaea can share a few conserved origins. Phylogenetic and genomic context analyses suggested that there is an original replication origin (oriC1) that was inherited from the ancestor of archaea, and several other origins were likely evolved and/or translocated within the haloarchaeal species.
Conclusion: This study provides detailed information about the diversity of multiple orc/cdc6-associated replication origins in haloarchaeal genomes, and provides novel insight into the evolution of multiple replication origins in Archaea.
Figures



Similar articles
-
Influence of Origin Recognition Complex Proteins on the Copy Numbers of Three Chromosomes in Haloferax volcanii.J Bacteriol. 2018 Aug 10;200(17):e00161-18. doi: 10.1128/JB.00161-18. Print 2018 Sep 1. J Bacteriol. 2018. PMID: 29941422 Free PMC article.
-
Association between the dynamics of multiple replication origins and the evolution of multireplicon genome architecture in haloarchaea.Genome Biol Evol. 2014 Oct 3;6(10):2799-810. doi: 10.1093/gbe/evu219. Genome Biol Evol. 2014. PMID: 25281843 Free PMC article.
-
An archaeal chromosomal autonomously replicating sequence element from an extreme halophile, Halobacterium sp. strain NRC-1.J Bacteriol. 2003 Oct;185(20):5959-66. doi: 10.1128/JB.185.20.5959-5966.2003. J Bacteriol. 2003. PMID: 14526006 Free PMC article.
-
Archaeal DNA Replication Origins and Recruitment of the MCM Replicative Helicase.Enzymes. 2016;39:169-90. doi: 10.1016/bs.enz.2016.03.002. Epub 2016 Apr 19. Enzymes. 2016. PMID: 27241930 Review.
-
Polyploidy in halophilic archaea: regulation, evolutionary advantages, and gene conversion.Biochem Soc Trans. 2019 Jun 28;47(3):933-944. doi: 10.1042/BST20190256. Epub 2019 Jun 12. Biochem Soc Trans. 2019. PMID: 31189733 Review.
Cited by
-
Origins of DNA replication.PLoS Genet. 2019 Sep 12;15(9):e1008320. doi: 10.1371/journal.pgen.1008320. eCollection 2019 Sep. PLoS Genet. 2019. PMID: 31513569 Free PMC article. Review.
-
Construction of Expression Shuttle Vectors for the Haloarchaeon Natrinema sp. J7 Based on Its Chromosomal Origins of Replication.Archaea. 2017 Mar 1;2017:4237079. doi: 10.1155/2017/4237079. eCollection 2017. Archaea. 2017. PMID: 28348508 Free PMC article.
-
Transcriptome analysis of Haloquadratum walsbyi: vanity is but the surface.BMC Genomics. 2017 Jul 3;18(1):510. doi: 10.1186/s12864-017-3892-2. BMC Genomics. 2017. PMID: 28673248 Free PMC article.
-
Haloferax volcanii-a model archaeon for studying DNA replication and repair.Open Biol. 2020 Dec;10(12):200293. doi: 10.1098/rsob.200293. Epub 2020 Dec 2. Open Biol. 2020. PMID: 33259746 Free PMC article. Review.
-
A virus-borne DNA damage signaling pathway controls the lysogeny-induction switch in a group of temperate pleolipoviruses.Nucleic Acids Res. 2023 Apr 24;51(7):3270-3287. doi: 10.1093/nar/gkad125. Nucleic Acids Res. 2023. PMID: 36864746 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

