Identification of suitable endogenous control genes for microRNA expression profiling of childhood medulloblastoma and human neural stem cells
- PMID: 22980291
- PMCID: PMC3531299
- DOI: 10.1186/1756-0500-5-507
Identification of suitable endogenous control genes for microRNA expression profiling of childhood medulloblastoma and human neural stem cells
Abstract
Background: Medulloblastoma (MB) is the most common type of malignant childhood brain tumour. Although deregulated microRNA (miRNA) expression has been linked to MB pathogenesis, the selection of appropriate candidate endogenous control (EC) reference genes for MB miRNA expression profiling studies has not been systematically addressed. In this study we utilised reverse transcriptase quantitative PCR (RT-qPCR) to identify the most appropriate EC reference genes for the accurate normalisation of miRNA expression data in primary human MB specimens and neural stem cells.
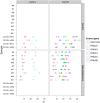
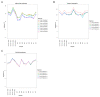
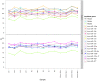
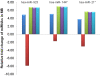
Results: Expression profiling of 662 miRNAs and six small nuclear/ nucleolar RNAs in primary human MB specimens, two CD133+ neural stem cell (NSC) populations and two CD133- neural progenitor cell (NPC) populations was performed using TaqMan low-density array (TLDA) cards. Minimal intra-card variability for candidate EC reference gene replicates was observed, however significant inter-card variability was identified between replicates present on both TLDA cards A and B. A panel of 18 potentially suitable EC reference genes was identified for the normalisation of miRNA expression on TLDA cards. These candidates were not significantly differentially expressed between CD133+ NSCs/ CD133- NPCs and primary MB specimens. Of the six sn/snoRNA EC reference genes recommended by the manufacturer, only RNU44 was uniformly expressed between primary MB specimens and CD133+ NSC/CD133- NPC populations (P = 0.709; FC = 1.02). The suitability of candidate EC reference genes was assessed using geNorm and NormFinder software, with hsa-miR-301a and hsa-miR-339-5p found to be the most uniformly expressed EC reference genes on TLDA card A and hsa-miR-425* and RNU24 for TLDA card B.
Conclusions: A panel of 18 potential EC reference genes that were not significantly differentially expressed between CD133+ NSCs/ CD133- NPCs and primary human MB specimens was identified. The top ranked EC reference genes described here should be validated in a larger cohort of specimens to verify their utility as controls for the normalisation of RT-qPCR data generated in MB miRNA expression studies. Importantly, inter-card variability observed between replicates of certain candidate EC reference genes has major implications for the accurate normalisation of miRNA expression data obtained using the miRNA TLDA platform.
Figures
References
-
- Giangaspero F, In: Medulloblastoma, in WHO Classification of Tumours of the Central Nervous System. Louis DN, editor. International Agency for Research on Cancer (IARC), Lyon; 2007. pp. 132–140.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

