Microprocessor, Setx, Xrn2, and Rrp6 co-operate to induce premature termination of transcription by RNAPII
- PMID: 22980978
- PMCID: PMC3595997
- DOI: 10.1016/j.cell.2012.08.004
Microprocessor, Setx, Xrn2, and Rrp6 co-operate to induce premature termination of transcription by RNAPII
Abstract
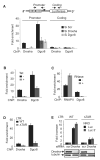
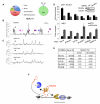
Transcription elongation is increasingly recognized as an important mechanism of gene regulation. Here, we show that microprocessor controls gene expression in an RNAi-independent manner. Microprocessor orchestrates the recruitment of termination factors Setx and Xrn2, and the 3'-5' exoribonuclease, Rrp6, to initiate RNAPII pausing and premature termination at the HIV-1 promoter through cleavage of the stem-loop RNA, TAR. Rrp6 further processes the cleavage product, which generates a small RNA that is required to mediate potent transcriptional repression and chromatin remodeling at the HIV-1 promoter. Using chromatin immunoprecipitation coupled to high-throughput sequencing (ChIP-seq), we identified cellular gene targets whose transcription is modulated by microprocessor. Our study reveals RNAPII pausing and premature termination mediated by the co-operative activity of ribonucleases, Drosha/Dgcr8, Xrn2, and Rrp6, as a regulatory mechanism of RNAPII-dependent transcription elongation.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures



References
-
- Bennasser Y, Yeung ML, Jeang KT. HIV-1 TAR RNA subverts RNA interference in transfected cells through sequestration of TAR RNA-binding protein, TRBP. J. Biol. Chem. 2006;281:27674–27678. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

