Direct regulation of GTP homeostasis by (p)ppGpp: a critical component of viability and stress resistance
- PMID: 22981860
- PMCID: PMC3483369
- DOI: 10.1016/j.molcel.2012.08.009
Direct regulation of GTP homeostasis by (p)ppGpp: a critical component of viability and stress resistance
Abstract
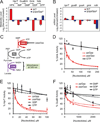
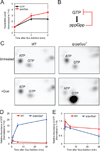
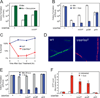
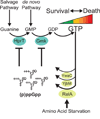
Cells constantly adjust their metabolism in response to environmental conditions, yet major mechanisms underlying survival remain poorly understood. We discover a posttranscriptional mechanism that integrates starvation response with GTP homeostasis to allow survival, enacted by the nucleotide (p)ppGpp, a key player in bacterial stress response and persistence. We reveal that (p)ppGpp activates global metabolic changes upon starvation, allowing survival by regulating GTP. Combining metabolomics with biochemical demonstrations, we find that (p)ppGpp directly inhibits the activities of multiple GTP biosynthesis enzymes. This inhibition results in robust and rapid GTP regulation in Bacillus subtilis, which we demonstrate is essential to maintaining GTP levels within a range that supports viability even in the absence of starvation. Correspondingly, without (p)ppGpp, gross GTP dysregulation occurs, revealing a vital housekeeping function of (p)ppGpp; in fact, loss of (p)ppGpp results in death from rising GTP, a severe and previously unknown consequence of GTP dysfunction.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures



References
-
- Artsimovitch I, Patlan V, Sekine S, Vassylyeva MN, Hosaka T, Ochi K, Yokoyama S, Vassylyev DG. Structural basis for transcription regulation by alarmone ppGpp. Cell. 2004;117:299–310. - PubMed
-
- Barker MM, Gaal T, Josaitis CA, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J Mol Biol. 2001;305:673–688. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

