Elements of ribosomal drug resistance and specificity
- PMID: 22981944
- PMCID: PMC5560274
- DOI: 10.1016/j.sbi.2012.07.016
Elements of ribosomal drug resistance and specificity
Abstract
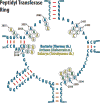
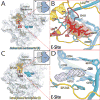
The structures of ribosomes in complex with inhibitors of translation have not only shed light on the interactions of antibiotics with the ribosome but also on the underlying mechanisms by which they interfere with the ribosome function. Several recent papers [1(•),2(••),3,4] have correlated the available ribosome structures with the wealth of biochemical data [5(•)]. In this review we shall focus on the lessons learned for drug specificity rather than presenting a comprehensive survey of the known structures of ribosome complexes with antibiotics.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures


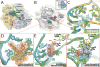
References
-
- Wilson DN. The A–Z of bacterial translation inhibitors. Critical Reviews in Biochemistry and Molecular Biology. 2009;44:393–433. An extensive review of the recent structures of inhibitors bound to the ribosome. - PubMed
-
- Sohmen D, Harms JM, Schlunzen F, Wilson DN. Enhanced SnapShot: Antibiotic inhibition of protein synthesis II. Cell. 2009;139:212–212 e211. An appealing graphical representation of the effects of many different classes of inhibitors on protein translation. - PubMed
-
- Poehlsgaard J, Douthwaite S. The bacterial ribosome as a target for antibiotics. Nat Rev Microbiol. 2005;3:870–881. - PubMed
-
- Kannan K, Mankin AS. Macrolide antibiotics in the ribosome exit tunnel: species-specific binding and action. Antimicrobial Therapeutics Reviews: Antibiotics That Target the Ribosome. 2011;1241:33–47. - PubMed
-
- Spahn CM, Prescott CD. Throwing a spanner in the works: antibiotics and the translation apparatus. J Mol Med (Berl) 1996;74:423–439. in combination with [1] and [41] represent most of the knowledge accumulated on inhibitors of protein translation over the past decades. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

