Bayesian statistical analysis of circadian oscillations in fibroblasts
- PMID: 22982138
- PMCID: PMC3478438
- DOI: 10.1016/j.jtbi.2012.08.038
Bayesian statistical analysis of circadian oscillations in fibroblasts
Abstract
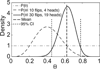
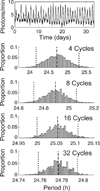
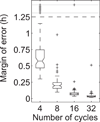
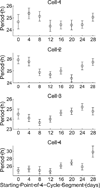
Precise determination of a noisy biological oscillator's period from limited experimental data can be challenging. The common practice is to calculate a single number (a point estimate) for the period of a particular time course. Uncertainty is inherent in any statistical estimator applied to noisy data, so our confidence in such point estimates depends on the quality and quantity of the data. Ideally, a period estimation method should both produce an accurate point estimate of the period and measure the uncertainty in that point estimate. A variety of period estimation methods are known, but few assess the uncertainty of the estimates, and a measure of uncertainty is rarely reported in the experimental literature. We compare the accuracy of point estimates using six common methods, only one of which can also produce uncertainty measures. We then illustrate the advantages of a new Bayesian method for estimating period, which outperforms the other six methods in accuracy of point estimates for simulated data and also provides a measure of uncertainty. We apply this method to analyze circadian oscillations of gene expression in individual mouse fibroblast cells and compute the number of cells and sampling duration required to reduce the uncertainty in period estimates to a desired level. This analysis indicates that, due to the stochastic variability of noisy intracellular oscillators, achieving a narrow margin of error can require an impractically large number of cells. In addition, we use a hierarchical model to determine the distribution of intrinsic cell periods, thereby separating the variability due to stochastic gene expression within each cell from the variability in period across the population of cells.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures





References
-
- Andrieu C, Doucet A. Joint Bayesian model selection and estimation of noisy sinusoids via reversible jump MCMC. IEEE Transactions on Signal Processing. 1999;47:2667–2676.
-
- Andrieu C, N DF, Doucet A, Jordan MI. An introduction to MCMC for machine learning. Machine Learning. 2003;50:5–43.
-
- Bretthorst GL. Bayesian Spectrum Analysis and Parameter Estimation (Lecture Notes in Statistics 48) Berlin: Springer; 1988.
-
- Chib S, Greenberg E. Understanding the Metropolis--Hastings algorithm. The American Statistician. 1995;49:327–335.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

