Structural analysis of the quaking homodimerization interface
- PMID: 22982292
- PMCID: PMC3472039
- DOI: 10.1016/j.jmb.2012.08.027
Structural analysis of the quaking homodimerization interface
Abstract
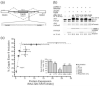
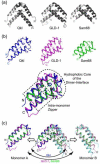
Quaking (QkI) is a prototypical member of the STAR (signal transducer and activator of RNA) protein family, which plays key roles in posttranscriptional gene regulation by controlling mRNA translation, stability and splicing. QkI-5 has been shown to regulate mRNA expression in the central nervous system, but little is known about its roles in other tissues. STAR proteins function as dimers and bind to bipartite RNA sequences; however, the structural and functional roles of homodimerization and heterodimerization are still unclear. Here, we present the crystal structure of the QkI dimerization domain, which adopts a similar stacked helix-turn-helix arrangement as its homologs GLD-1 (germ line development defective-1) and Sam68 (Src-associated protein during mitosis, 68kDa) but differs by an additional helix inserted in the dimer interface. Variability of the dimer interface residues likely ensures selective homodimerization by preventing association with non-cognate STAR family proteins in the cell. Mutations that inhibit dimerization also significantly impair RNA binding in vitro, alter QkI-5 protein levels and impair QkI function in a splicing assay in vivo. Together, our results indicate that a functional Qua1 homodimerization domain is required for QkI-5 function in mammalian cells.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures



References
-
- Artzt K, Wu JI. STAR trek: An introduction to STAR family proteins and review of quaking (QKI) Adv Exp Med Biol. 2010;693:1–24. - PubMed
-
- Vernet C, Artzt K. STAR, a gene family involved in signal transduction and activation of RNA. Trends Genet. 1997;13:479–84. - PubMed
-
- Lobbardi R, Lambert G, Zhao J, Geisler R, Kim HR, Rosa FM. Fine-tuning of Hh signaling by the RNA-binding protein Quaking to control muscle development. Development. 2011;138:1783–94. - PubMed
-
- Noveroske JK, Lai L, Gaussin V, Northrop JL, Nakamura H, Hirschi KK, Justice MJ. Quaking is essential for blood vessel development. Genesis. 2002;32:218–30. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

