The cytochrome b5 dependent C-5(6) sterol desaturase DES5A from the endoplasmic reticulum of Tetrahymena thermophila complements ergosterol biosynthesis mutants in Saccharomyces cerevisiae
- PMID: 22982564
- PMCID: PMC3501532
- DOI: 10.1016/j.steroids.2012.08.015
The cytochrome b5 dependent C-5(6) sterol desaturase DES5A from the endoplasmic reticulum of Tetrahymena thermophila complements ergosterol biosynthesis mutants in Saccharomyces cerevisiae
Abstract
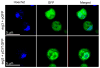
Tetrahymena thermophila is a free-living ciliate with no exogenous sterol requirement. However, it can perform several modifications on externally added sterols including desaturation at C5(6), C7(8), and C22(23). Sterol desaturases in Tetrahymena are microsomal enzymes that require Cyt b(5), Cyt b(5) reductase, oxygen, and reduced NAD(P)H for their activity, and some of the genes encoding these functions have recently been identified. The DES5A gene encodes a C-5(6) sterol desaturase, as shown by gene knockout in Tetrahymena. To confirm and extend that result, and to develop new approaches to gene characterization in Tetrahymena, we have now, expressed DES5A in Saccharomyces cerevisiae. The DES5A gene was codon optimized and expressed in a yeast mutant, erg3Δ, which is disrupted for the gene encoding the S. cerevisiae C-5(6) sterol desaturase ERG3. The complemented strain was able to accumulate 74% of the wild type level of ergosterol, and also lost the hypersensitivity to cycloheximide associated with the lack of ERG3 function. C-5(6) sterol desaturases are expected to function at the endoplasmic reticulum. Consistent with this, a GFP-tagged copy of Des5Ap was localized to the endoplasmic reticulum in both Tetrahymena and yeast. This work shows for the first time that both function and localization are conserved for a microsomal enzyme between ciliates and fungi, notwithstanding the enormous evolutionary distance between these lineages. The results suggest that heterologous expression of ciliate genes in S. cerevisiae provides a useful tool for the characterization of genes in Tetrahymena, including genes encoding membrane protein complexes.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures




References
-
- Volkman JK. Sterols in microorganisms. Appl Microbiol Biotechnol. 200;60:495–506. - PubMed
-
- Ikekawa N, Morisaki M, Fujimoto Y. Sterol metabolism in insects: dealkylation of phytosterol to cholesterol. Acc Chem Res. 1993;26:139–46.
-
- Thompson GA, Bamberry RJ, Nozawa Y. Further Studies of the Lipid Coposition and Biochemical Properties of Tetrahymena pyriformis Membrane Systems. Biochemistry. 1971;10:4441–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

