Investigating bacterial-animal symbioses with light sheet microscopy
- PMID: 22983029
- PMCID: PMC3952068
- DOI: 10.1086/BBLv223n1p7
Investigating bacterial-animal symbioses with light sheet microscopy
Abstract
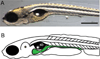
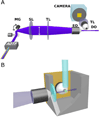
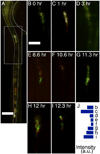
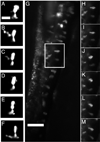
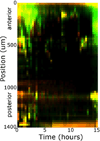
Microbial colonization of the digestive tract is a crucial event in vertebrate development, required for maturation of host immunity and establishment of normal digestive physiology. Advances in genomic, proteomic, and metabolomic technologies are providing a more detailed picture of the constituents of the intestinal habitat, but these approaches lack the spatial and temporal resolution needed to characterize the assembly and dynamics of microbial communities in this complex environment. We report the use of light sheet microscopy to provide high-resolution imaging of bacterial colonization of the intestine of Danio rerio, the zebrafish. The method allows us to characterize bacterial population dynamics across the entire organ and the behaviors of individual bacterial and host cells throughout the colonization process. The large four-dimensional data sets generated by these imaging approaches require new strategies for image analysis. When integrated with other "omics" data sets, information about the spatial and temporal dynamics of microbial cells within the vertebrate intestine will provide new mechanistic insights into how microbial communities assemble and function within hosts.
Figures

References
-
- Altarriba M, Merino S, Gavin R, Canals R, Rabaan A, Shaw JG, Tomas JM. A polar flagella operon (flg) of Aeromonas hydrophila contains genes required for lateral flagella expression. Microb Pathog. 2003;34:249–259. - PubMed
-
- Bates JM, Mittge E, Kuhlman J, Baden KN, Cheesman SE, Guillemin K. Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Dev Biol. 2006;297:374–386. - PubMed
-
- Bjornstad ON, Ims RA, Lambin X. Spatial population dynamics: analyzing patterns and processes of population synchrony. Trends Ecol Evol. 1999;14:427–432. - PubMed
-
- Boettcher KJ, Ruby EG, McFall-Ngai MJ. Bioluminescence in the symbiotic squid Euprymna scolopes is controlled by a daily biological rhythm. Journal of Comparative Physiology A Sensory Neural and Behavioral Physiology. 1996;179:65–73.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

