Maternal allergen exposure reprograms the developmental lung transcriptome in atopic and normoresponsive rat pups
- PMID: 22983352
- PMCID: PMC3517678
- DOI: 10.1152/ajplung.00179.2012
Maternal allergen exposure reprograms the developmental lung transcriptome in atopic and normoresponsive rat pups
Abstract
The "fetal origins hypothesis" argued that physiological changes consequent to in utero exposures ultimately contribute to disease susceptibility in later life. The dramatic increase in asthma prevalence is attributed to early exposures acting on preexisting asthma-susceptible genotypes. We showed previously that distinct transcriptome signatures distinguish the developmental respiratory phenotype of atopic (Brown Norway, BN) and normoresponsive (Lewis) rats. We aimed to determine whether maternal allergen exposure would influence asthma pathogenesis by reprogramming primary patterns of developmental lung gene expression. Postnatal offspring of dams sensitized to ovalbumin before mating and challenged during pregnancy were assessed for lung function, inflammatory biomarkers, and respiratory gene expression. Although maternal ovalbumin exposure resulted in characteristic features of an allergic response (bronchoalveolar lavage neutrophils, IgE, methacholine-induced lung resistance) in offspring of both strains, substantial strain-specific differences were observed in respiratory gene expression. Of 799 probes representing the top 5% of transcriptomic variation, only 112 (14%) were affected in both strains. Strain-specific gene signatures also exhibited marked differences in enrichment for gene ontologies, with immune regulation and cell proliferation being prominent in the BN strain, cell cycle and microtubule assembly gene sets in the Lewis strain. Multiple ovalbumin-specific probes in both strains were also differentially expressed in lymphoblastoid cell lines from human asthmatic vs. nonasthmatic sibling pairs. Our data point to the existence of distinct, genetically programmed responses to maternal exposures in developing lung. These different response patterns, if recapitulated in human fetal development, can contribute to long-term pulmonary health including interindividual susceptibility to asthma.
Figures


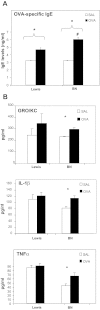
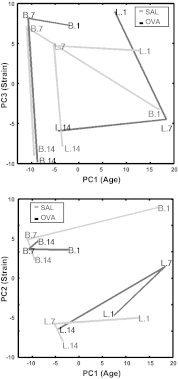
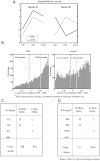



References
-
- Baraldi E, Filippone M. Chronic lung disease after premature birth. N Engl J Med 357: 1946– 1955, 2007 - PubMed
-
- Barker D. Fetal and Neonatal Origins of Adult Disease. London: BMJ, 1992
-
- Bellofiore S, Martin JG. Antigen challenge of sensitized rats increases airway responsiveness to methacholine. J Appl Physiol 65: 1642– 1646, 1988 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

