Stable transfection of the diplomonad parasite Spironucleus salmonicida
- PMID: 22983987
- PMCID: PMC3486028
- DOI: 10.1128/EC.00179-12
Stable transfection of the diplomonad parasite Spironucleus salmonicida
Abstract
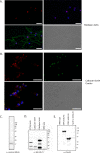
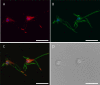
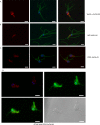
Eukaryotic microbes are highly diverse, and many lineages remain poorly studied. One such lineage, the diplomonads, a group of binucleate heterotrophic flagellates, has been studied mainly due to the impact of Giardia intestinalis, an intestinal, diarrhea-causing parasite in humans and animals. Here we describe the development of a stable transfection system for use in Spironucleus salmonicida, a diplomonad that causes systemic spironucleosis in salmonid fish. We designed vectors in cassette format carrying epitope tags for localization (3×HA [where HA is hemagglutinin], 2× Escherichia coli OmpF linker and mouse langerin fusion sequence [2×OLLAS], 3×MYC) and purification of proteins (2× Strep-Tag II-FLAG tandem-affinity purification tag or streptavidin binding peptide-glutathione S-transferase [SBP-GST]) under the control of native or constitutive promoters. Three selectable gene markers, puromycin acetyltransferase (pac), blasticidin S-deaminase (bsr), and neomycin phosphotransferase (nptII), were successfully applied for the generation of stable transfectants. Site-specific integration on the S. salmonicida chromosome was shown to be possible using the bsr resistance gene. We epitope tagged six proteins and confirmed their expression by Western blotting. Next, we demonstrated the utility of these vectors by recording the subcellular localizations of the six proteins by laser scanning confocal microscopy. Finally, we described the creation of an S. salmonicida double transfectant suitable for colocalization studies. The transfection system described herein and the imminent completion of the S. salmonicida genome will make it possible to use comparative genomics as an investigative tool to explore specific, as well as general, diplomonad traits, benefiting research on both Giardia and Spironucleus.
Figures

Similar articles
-
Stable transformation of an episomal protein-tagging shuttle vector in the piscine diplomonad Spironucleus vortens.BMC Microbiol. 2008 Apr 29;8:71. doi: 10.1186/1471-2180-8-71. BMC Microbiol. 2008. PMID: 18445284 Free PMC article.
-
Large genomic differences between the morphologically indistinguishable diplomonads Spironucleus barkhanus and Spironucleus salmonicida.BMC Genomics. 2010 Apr 21;11:258. doi: 10.1186/1471-2164-11-258. BMC Genomics. 2010. PMID: 20409319 Free PMC article.
-
The genome of Spironucleus salmonicida highlights a fish pathogen adapted to fluctuating environments.PLoS Genet. 2014 Feb 6;10(2):e1004053. doi: 10.1371/journal.pgen.1004053. eCollection 2014 Feb. PLoS Genet. 2014. PMID: 24516394 Free PMC article.
-
Comparative biochemistry of Giardia, Hexamita and Spironucleus: Enigmatic diplomonads.Mol Biochem Parasitol. 2014 Oct;197(1-2):43-9. doi: 10.1016/j.molbiopara.2014.10.002. Epub 2014 Oct 16. Mol Biochem Parasitol. 2014. PMID: 25448769 Review.
-
Several affinity tags commonly used in chromatographic purification.J Anal Methods Chem. 2013;2013:581093. doi: 10.1155/2013/581093. Epub 2013 Dec 26. J Anal Methods Chem. 2013. PMID: 24490106 Free PMC article. Review.
Cited by
-
Comparative Cell Biology and Evolution of Annexins in Diplomonads.mSphere. 2016 Mar 23;1(2):e00032-15. doi: 10.1128/mSphere.00032-15. eCollection 2016 Mar-Apr. mSphere. 2016. PMID: 27303715 Free PMC article.
-
Combined nanometric and phylogenetic analysis of unique endocytic compartments in Giardia lamblia sheds light on the evolution of endocytosis in Metamonada.BMC Biol. 2022 Sep 21;20(1):206. doi: 10.1186/s12915-022-01402-3. BMC Biol. 2022. PMID: 36127707 Free PMC article.
-
Oxygen induces the expression of invasion and stress response genes in the anaerobic salmon parasite Spironucleus salmonicida.BMC Biol. 2019 Mar 1;17(1):19. doi: 10.1186/s12915-019-0634-8. BMC Biol. 2019. PMID: 30823887 Free PMC article.
-
Proximity Staining Using Enzymatic Protein Tagging in Diplomonads.mSphere. 2019 Mar 20;4(2):e00153-19. doi: 10.1128/mSphereDirect.00153-19. mSphere. 2019. PMID: 30894436 Free PMC article.
-
Cyst-Wall-Protein-1 is fundamental for Golgi-like organelle neogenesis and cyst-wall biosynthesis in Giardia lamblia.Nat Commun. 2016 Dec 15;7:13859. doi: 10.1038/ncomms13859. Nat Commun. 2016. PMID: 27976675 Free PMC article.
References
-
- Andersson JO, Sjogren AM, Davis LA, Embley TM, Roger AJ. 2003. Phylogenetic analyses of diplomonad genes reveal frequent lateral gene transfers affecting eukaryotes. Curr. Biol. 13:94–104 - PubMed
-
- Andersson JO, et al. 2007. A genomic survey of the fish parasite Spironucleus salmonicida indicates genomic plasticity among diplomonads and significant lateral gene transfer in eukaryote genome evolution. BMC Genomics 8:51 doi:10.1186/1471-2164-8-51 - DOI - PMC - PubMed
-
- Benne R. 1994. RNA editing in trypanosomes. Eur. J. Biochem. 221:9–23 - PubMed
-
- Brinkmann H, Philippe H. 2007. The diversity of eukaryotes and the root of the eukaryotic tree. Adv. Exp. Med. Biol. 607:20–37 - PubMed
-
- Correa G, Morgado-Diaz JA, Benchimol M. 2004. Centrin in Giardia lamblia—ultrastructural localization. FEMS Microbiol. Lett. 233:91–96 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

