Regulation and functional significance of autophagy in respiratory cell biology and disease
- PMID: 22984088
- PMCID: PMC3547078
- DOI: 10.1165/rcmb.2012-0282TR
Regulation and functional significance of autophagy in respiratory cell biology and disease
Abstract
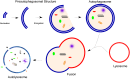
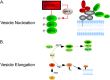
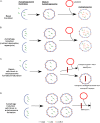
Autophagy is a homeostatic process common to all eukaryotic cells that serves to degrade intracellular components. Among three classes of autophagy, macroautophagy is best understood, and is the subject of this Review. The function of autophagy is multifaceted, and includes removal of long-lived proteins and damaged or unneeded organelles, recycling of intracellular components for nutrients, and defense against pathogens. This process has been extensively studied in yeast, and understanding of its functional significance in human disease is also increasing. This Review explores the basic machinery and regulation of autophagy in mammalian systems, methods employed to measure autophagic activity, and then focuses on recent discoveries about the functional significance of autophagy in respiratory diseases, including chronic obstructive pulmonary disease, cystic fibrosis, tuberculosis, idiopathic pulmonary fibrosis, pulmonary arterial hypertension, acute lung injury, and lymphangioleiomyomatosis.
Figures
References
-
- Tolkovsky AM. Mitophagy. Biochim Biophys Acta 2009;1793:1508–1515 - PubMed
-
- Bernales S, Schuck S, Walter P. ER-phagy: selective autophagy of the endoplasmic reticulum. Autophagy 2007;3:285–287 - PubMed
-
- Sakai Y, Oku M, van der Klei IJ, Kiel JA. Pexophagy: autophagic degradation of peroxisomes. Biochim Biophys Acta 2006;1763:1767–1775 - PubMed
-
- Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the UBP3P/BRE5P ubiquitin protease. Nat Cell Biol 2008;10:602–610 - PubMed
-
- Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell 2005;120:159–162 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

