Bacillus subtilis hlpB encodes a conserved stand-alone HNH nuclease-like protein that is essential for viability unless the hlpB deletion is accompanied by the deletion of genes encoding the AddAB DNA repair complex
- PMID: 22984257
- PMCID: PMC3486360
- DOI: 10.1128/JB.05283-11
Bacillus subtilis hlpB encodes a conserved stand-alone HNH nuclease-like protein that is essential for viability unless the hlpB deletion is accompanied by the deletion of genes encoding the AddAB DNA repair complex
Abstract
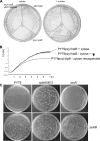
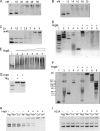
The HNH domain is found in many different proteins in all phylogenetic kingdoms and in many cases confers nuclease activity. We have found that the Bacillus subtilis hlpB (yisB) gene encodes a stand-alone HNH domain, homologs of which are present in several bacterial genomes. We show that the protein we term HlpB is essential for viability. The depletion of HlpB leads to growth arrest and to the generation of cells containing a single, decondensed nucleoid. This apparent condensation-segregation defect was cured by additional hlpB copies in trans. Purified HlpB showed cooperative binding to a variety of double-stranded and single-stranded DNA sequences, depending on the presence of zinc, nickel, or cobalt ions. Binding of HlpB was also influenced by pH and different metals, reminiscent of HNH domains. Lethality of the hlpB deletion was relieved in the absence of addA and of addAB, two genes encoding proteins forming a RecBCD-like end resection complex, but not of recJ, which is responsible for a second end-resectioning avenue. Like AddA-green fluorescent protein (AddA-GFP), functional HlpB-YFP or HlpB-FlAsH fusions were present throughout the cytosol in growing B. subtilis cells. Upon induction of DNA damage, HlpB-FlAsH formed a single focus on the nucleoid in a subset of cells, many of which colocalized with the replication machinery. Our data suggest that HlpB plays a role in DNA repair by rescuing AddAB-mediated recombination intermediates in B. subtilis and possibly also in many other bacteria.
Figures





Similar articles
-
RecA Is Required for the Assembly of RecN into DNA Repair Complexes on the Nucleoid.J Bacteriol. 2021 Sep 23;203(20):e0024021. doi: 10.1128/JB.00240-21. Epub 2021 Aug 2. J Bacteriol. 2021. PMID: 34339298 Free PMC article.
-
Bacillus subtilis SbcC protein plays an important role in DNA inter-strand cross-link repair.BMC Mol Biol. 2006 Jun 16;7:20. doi: 10.1186/1471-2199-7-20. BMC Mol Biol. 2006. PMID: 16780573 Free PMC article.
-
Recruitment of Bacillus subtilis RecN to DNA double-strand breaks in the absence of DNA end processing.J Bacteriol. 2006 Jan;188(2):353-60. doi: 10.1128/JB.188.2.353-360.2006. J Bacteriol. 2006. PMID: 16385024 Free PMC article.
-
Review of DNA repair enzymes in bacteria: With a major focus on AddAB and RecBCD.DNA Repair (Amst). 2022 Oct;118:103389. doi: 10.1016/j.dnarep.2022.103389. Epub 2022 Aug 24. DNA Repair (Amst). 2022. PMID: 36030574 Review.
-
Double-strand break repair in bacteria: a view from Bacillus subtilis.FEMS Microbiol Rev. 2011 Nov;35(6):1055-81. doi: 10.1111/j.1574-6976.2011.00272.x. Epub 2011 May 18. FEMS Microbiol Rev. 2011. PMID: 21517913 Review.
Cited by
-
Systematic classification of the His-Me finger superfamily.Nucleic Acids Res. 2017 Nov 16;45(20):11479-11494. doi: 10.1093/nar/gkx924. Nucleic Acids Res. 2017. PMID: 29040665 Free PMC article.
-
Structural and functional characterization of deep-sea thermophilic bacteriophage GVE2 HNH endonuclease.Sci Rep. 2017 Feb 13;7:42542. doi: 10.1038/srep42542. Sci Rep. 2017. PMID: 28211904 Free PMC article.
-
Gradients in gene essentiality reshape antibacterial research.FEMS Microbiol Rev. 2022 May 6;46(3):fuac005. doi: 10.1093/femsre/fuac005. FEMS Microbiol Rev. 2022. PMID: 35104846 Free PMC article. Review.
-
Visual Evidence for the Recruitment of Four Enzymes with RNase Activity to the Bacillus subtilis Replication Forks.Cells. 2024 Aug 20;13(16):1381. doi: 10.3390/cells13161381. Cells. 2024. PMID: 39195267 Free PMC article.
-
Structure determination and biochemical characterization of a putative HNH endonuclease from Geobacter metallireducens GS-15.PLoS One. 2013 Sep 6;8(9):e72114. doi: 10.1371/journal.pone.0072114. eCollection 2013. PLoS One. 2013. PMID: 24039739 Free PMC article.
References
-
- Beall B, Lutkenhaus J. 1991. FtsZ in Bacillus subtilis is required for vegetative septation and for asymmetric septation during sporulation. Genes Dev. 5:447–455 - PubMed
-
- Bujnicki JM, Radlinska M, Rychlewski L. 2000. Atomic model of the 5-methylcytosine-specific restriction enzyme McrA reveals an atypical zinc finger and structural similarity to betabetaalphaMe endonucleases. Mol. Microbiol. 37:1280–1281 - PubMed
-
- Chédin F, Handa N, Dillingham MS, Kowalczykowski SC. 2006. The AddAB helicase/nuclease forms a stable complex with its cognate chi sequence during translocation. J. Biol. Chem. 281:18610–18617 - PubMed
-
- Cheng YS, Hsia KC, Doudeva LG, Chak KF, Yuan HS. 2002. The crystal structure of the nuclease domain of colicin E7 suggests a mechanism for binding to double-stranded DNA by the H-N-H endonucleases. J. Mol. Biol. 324:227–236 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

