Polar/Ionizable residues in transmembrane segments: effects on helix-helix packing
- PMID: 22984481
- PMCID: PMC3440369
- DOI: 10.1371/journal.pone.0044263
Polar/Ionizable residues in transmembrane segments: effects on helix-helix packing
Abstract
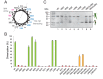
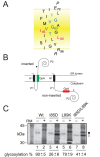
The vast majority of membrane proteins are anchored to biological membranes through hydrophobic α-helices. Sequence analysis of high-resolution membrane protein structures show that ionizable amino acid residues are present in transmembrane (TM) helices, often with a functional and/or structural role. Here, using as scaffold the hydrophobic TM domain of the model membrane protein glycophorin A (GpA), we address the consequences of replacing specific residues by ionizable amino acids on TM helix insertion and packing, both in detergent micelles and in biological membranes. Our findings demonstrate that ionizable residues are stably inserted in hydrophobic environments, and tolerated in the dimerization process when oriented toward the lipid face, emphasizing the complexity of protein-lipid interactions in biological membranes.
Conflict of interest statement
Figures



References
-
- Popot JL, Engelman DM (1990) Membrane protein folding and oligomerization - The 2-stage model. Biochemistry 29: 4031–4037. - PubMed
-
- Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, et al. (2005) Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature 433: 377–381. - PubMed
-
- Baeza-Delgado C, Marti-Renom MA, Mingarro I (accepted) Structure-based statistical analysis of transmembrane segments. Eur Biophys J DOI 10.1007/s00249-012-0813-9. - PubMed
-
- Hessa T, Meindl-Beinker NM, Bernsel A, Kim H, Sato Y, et al. (2007) Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature 450: 1026–1030. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

