Gene duplicability-connectivity-complexity across organisms and a neutral evolutionary explanation
- PMID: 22984517
- PMCID: PMC3439388
- DOI: 10.1371/journal.pone.0044491
Gene duplicability-connectivity-complexity across organisms and a neutral evolutionary explanation
Abstract
Gene duplication has long been acknowledged by biologists as a major evolutionary force shaping genomic architectures and characteristics across the Tree of Life. Major research has been conducting on elucidating the fate of duplicated genes in a variety of organisms, as well as factors that affect a gene's duplicability--that is, the tendency of certain genes to retain more duplicates than others. In particular, two studies have looked at the correlation between gene duplicability and its degree in a protein-protein interaction network in yeast, mouse, and human, and another has looked at the correlation between gene duplicability and its complexity (length, number of domains, etc.) in yeast. In this paper, we extend these studies to six species, and two trends emerge. There is an increase in the duplicability-connectivity correlation that agrees with the increase in the genome size as well as the phylogenetic relationship of the species. Further, the duplicability-complexity correlation seems to be constant across the species. We argue that the observed correlations can be explained by neutral evolutionary forces acting on the genomic regions containing the genes. For the duplicability-connectivity correlation, we show through simulations that an increasing trend can be obtained by adjusting parameters to approximate genomic characteristics of the respective species. Our results call for more research into factors, adaptive and non-adaptive alike, that determine a gene's duplicability.
Conflict of interest statement
Figures
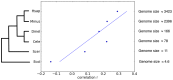
 ) between gene duplicability and gene connectivity for six species: H. sapien (Hsap), M. musculus (Mmus), D. melanogaster (Dmel), C. elegans (Cele), S. cerevisiae (Scer), and E. coli (Ecol). The evolutionary relationship of the species is based in part on . Genome size (in Mbp) information for all species, except E. coli, were obtained from the Animal Genome Size Database and the Fungal Genome Database.
) between gene duplicability and gene connectivity for six species: H. sapien (Hsap), M. musculus (Mmus), D. melanogaster (Dmel), C. elegans (Cele), S. cerevisiae (Scer), and E. coli (Ecol). The evolutionary relationship of the species is based in part on . Genome size (in Mbp) information for all species, except E. coli, were obtained from the Animal Genome Size Database and the Fungal Genome Database.
 ) between gene duplicability and gene connectivity for different settings under the subfunctionalization model (model Ib in [5]) and the neofunctionalization model (model IIc in [5]). The parameter values in each of the four settings are given in Table 2.
) between gene duplicability and gene connectivity for different settings under the subfunctionalization model (model Ib in [5]) and the neofunctionalization model (model IIc in [5]). The parameter values in each of the four settings are given in Table 2.Similar articles
-
Protein connectivity and protein complexity promotes human gene duplicability in a mutually exclusive manner.DNA Res. 2010 Oct;17(5):261-70. doi: 10.1093/dnares/dsq019. Epub 2010 Sep 9. DNA Res. 2010. PMID: 20829394 Free PMC article.
-
Relationship between gene duplicability and diversifiability in the topology of biochemical networks.BMC Genomics. 2014 Jul 8;15(1):577. doi: 10.1186/1471-2164-15-577. BMC Genomics. 2014. PMID: 25005725 Free PMC article.
-
Protein function, connectivity, and duplicability in yeast.Mol Biol Evol. 2006 Jan;23(1):30-9. doi: 10.1093/molbev/msi249. Epub 2005 Aug 24. Mol Biol Evol. 2006. PMID: 16120800
-
Evolutionary relationships between Saccharomyces cerevisiae and other fungal species as determined from genome comparisons.Rev Iberoam Micol. 2005 Dec;22(4):217-22. doi: 10.1016/s1130-1406(05)70046-2. Rev Iberoam Micol. 2005. PMID: 16499414 Review.
-
Birth and death of duplicated genes in completely sequenced eukaryotes.Trends Genet. 2001 May;17(5):237-9. doi: 10.1016/s0168-9525(01)02243-0. Trends Genet. 2001. PMID: 11335019 Review.
Cited by
-
Functional compensation of mouse duplicates by their paralogs expressed in the same tissues.Genome Biol Evol. 2022 Aug 10;14(8):evac126. doi: 10.1093/gbe/evac126. Online ahead of print. Genome Biol Evol. 2022. PMID: 35945673 Free PMC article.
-
Evolution after whole-genome duplication: a network perspective.G3 (Bethesda). 2013 Nov 6;3(11):2049-57. doi: 10.1534/g3.113.008458. G3 (Bethesda). 2013. PMID: 24048644 Free PMC article.
-
Extensive local gene duplication and functional divergence among paralogs in Atlantic salmon.Genome Biol Evol. 2014 Jun 19;6(7):1790-805. doi: 10.1093/gbe/evu131. Genome Biol Evol. 2014. PMID: 24951567 Free PMC article.
-
Dependency Between Protein-Protein Interactions and Protein Variability and Evolutionary Rates in Vertebrates: Observed Relationships and Stochastic Modeling.J Mol Evol. 2019 Jul;87(4-6):184-198. doi: 10.1007/s00239-019-09899-z. Epub 2019 Jul 13. J Mol Evol. 2019. PMID: 31302723 Free PMC article.
References
-
- Ohno S (1970) Evolution by gene duplication. Springer-Verlag.
-
- Dittmar K, Liberies D (2010) Evolution after gene duplication. Wiley
-
- Yamada T, Bork P (2009) Evolution of biomolecular networks: lessons from metabolic and protein interactions. Nature Reviews-Molecular Cell Biology 10: 791–803. - PubMed
-
- Roth C, Rastogi S, Arvestad L, Dittmar K, Light S, et al. (2007) Evolution after gene duplication: models, mechanisms, sequences, systems, and organisms. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution 308B: 58–73. - PubMed
-
- Innan H, Kondrashov F (2010) The evolution of gene duplications: classifying and distinguishing between models. Nature Reviews Genetics 11: 97–108. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

