Phylogenetic analysis reveals common antimicrobial resistant Campylobacter coli population in antimicrobial-free (ABF) and commercial swine systems
- PMID: 22984540
- PMCID: PMC3440321
- DOI: 10.1371/journal.pone.0044662
Phylogenetic analysis reveals common antimicrobial resistant Campylobacter coli population in antimicrobial-free (ABF) and commercial swine systems
Abstract
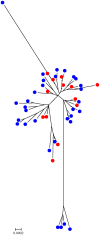
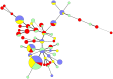
The objective of this study was to compare the population biology of antimicrobial resistant (AR) Campylobacter coli isolated from swine reared in the conventional and antimicrobial-free (ABF) swine production systems at farm, slaughter and environment. A total of 200 C. coli isolates selected from fecal, environmental, and carcass samples of ABF (n = 100) and conventional (n = 100) swine production systems were typed by multilocus sequence typing (MLST). Sequence data from seven housekeeping genes was analyzed for the identification of allelic profiles, sequence types (STs) and clonal complex determination. Phylogenetic trees were generated to establish the relationships between the genotyped isolates. A total of 51 STs were detected including two novel alleles (glnA 424 and glyA 464) and 14 novel STs reported for the first time. The majority of the C. coli isolates belonged to ST-854 (ABF: 31, conventional: 17), and were grouped in clonal complex ST-828 (ABF: 68%, conventional: 66%). The mean genetic diversity (H) for the ABF (0.3963+/-0.0806) and conventional (0.4655+/-0.0714) systems were similar. The index of association (I(A)(S)) for the ABF (I(A)(S)= 0.1513) and conventional (I(A)(S) = 0.0991) C. coli populations were close to linkage equilibrium, indicative of a freely recombining population. Identical STs were detected between the pigs and their environment both at farm and slaughter. A minimum spanning tree revealed the close clustering of C. coli STs that originated from swine and carcass with those from the environment. In conclusion, our study reveals a genotypic diverse C. coli population that shares a common ancestry in the conventional and ABF swine production systems. This could potentially explain the high prevalence of antimicrobial resistant C. coli in the ABF system in the absence of antimicrobial selection pressure.
Conflict of interest statement
Figures
References
-
- Hermans D, Pasmans F, Messens W, Martel A, Van Immerseel F, et al. (2012) Poultry as a host for the zoonotic pathogen Campylobacter jejuni . Vector Borne Zoonotic Dis 12: 89–98. - PubMed
-
- Grove-White DH, Leatherbarrow AJ, Cripps PJ, Diggle PJ, French NP (2010) Temporal and farm-management-associated variation in the faecal-pat prevalence of Campylobacter jejuni in ruminants. Epidemiol Infect 138: 549–558. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

