Mining gene expression signature for the detection of pre-malignant melanocytes and early melanomas with risk for metastasis
- PMID: 22984562
- PMCID: PMC3439384
- DOI: 10.1371/journal.pone.0044800
Mining gene expression signature for the detection of pre-malignant melanocytes and early melanomas with risk for metastasis
Abstract
Background: Metastatic melanoma is a highly aggressive skin cancer and currently resistant to systemic therapy. Melanomas may involve genetic, epigenetic and metabolic abnormalities. Evidence is emerging that epigenetic changes might play a significant role in tumor cell plasticity and metastatic phenotype of melanoma cells.
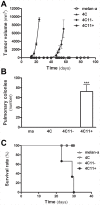
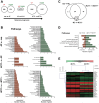
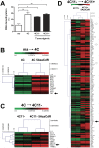
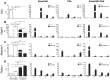
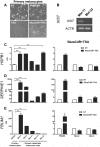
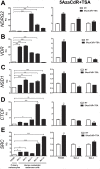
Principal findings: In this study, we developed a systematic approach to identify genes implicated in melanoma progression. To do this, we used the Affymetrix GeneChip Arrays to screen 34,000 mouse transcripts in melan-a melanocytes, 4C pre-malignant melanocytes, 4C11- non-metastatic and 4C11+ metastatic melanoma cell lines. The genome-wide association studies revealed pathways commonly over-represented in the transition from immortalized to pre-malignant stage, and under-represented in the transition from non-metastatic to metastatic stage. Additionally, the treatment of cells with 10 µM 5-aza-2'-deoxycytidine (5AzaCdR) for 48 hours allowed us to identify genes differentially re-expressed at specific stages of melan-a malignant transformation. Treatment of human primary melanocytes with the demethylating agent 5AzaCdR in combination to the histone deacetylase inhibitor Trichostatin A (TSA) revealed changes on melanocyte morphology and gene expression which could be an indicator of epigenetic flexibility in normal melanocytes. Moreover, changes on gene expression recognized by affecting the melanocyte biology (NDRG2 and VDR), phenotype of metastatic melanoma cells (HSPB1 and SERPINE1) and response to cancer therapy (CTCF, NSD1 and SRC) were found when Mel-2 and/or Mel-3-derived patient metastases were exposed to 5AzaCdR plus TSA treatment. Hierarchical clustering and network analyses in a panel of five patient-derived metastatic melanoma cells showed gene interactions that have never been described in melanomas.
Significance: Despite the heterogeneity observed in melanomas, this study demonstrates the utility of our murine melanoma progression model to identify molecular markers commonly perturbed in metastasis. Additionally, the novel gene expression signature identified here may be useful in the future into a model more closely related to translational research.
Conflict of interest statement
Figures

Similar articles
-
Epigenetic reprogramming as a key contributor to melanocyte malignant transformation.Epigenetics. 2011 Apr;6(4):450-64. doi: 10.4161/epi.6.4.14917. Epub 2011 Apr 1. Epigenetics. 2011. PMID: 21343701 Free PMC article.
-
Gene co-expression and histone modification signatures are associated with melanoma progression, epithelial-to-mesenchymal transition, and metastasis.Clin Epigenetics. 2020 Aug 24;12(1):127. doi: 10.1186/s13148-020-00910-9. Clin Epigenetics. 2020. PMID: 32831131 Free PMC article.
-
Timp1 interacts with beta-1 integrin and CD63 along melanoma genesis and confers anoikis resistance by activating PI3-K signaling pathway independently of Akt phosphorylation.Mol Cancer. 2013 Mar 25;12:22. doi: 10.1186/1476-4598-12-22. Mol Cancer. 2013. PMID: 23522389 Free PMC article.
-
Biology of tumor progression in human melanocytes.Lab Invest. 1987 May;56(5):461-74. Lab Invest. 1987. PMID: 3553733 Review.
-
Molecular biology of normal melanocytes and melanoma cells.J Clin Pathol. 2013 Aug;66(8):644-8. doi: 10.1136/jclinpath-2013-201471. Epub 2013 Mar 23. J Clin Pathol. 2013. PMID: 23526597 Review.
Cited by
-
Genome-wide promoter methylation profiling in a cellular model of melanoma progression reveals markers of malignancy and metastasis that predict melanoma survival.Clin Epigenetics. 2022 May 23;14(1):68. doi: 10.1186/s13148-022-01291-x. Clin Epigenetics. 2022. PMID: 35606887 Free PMC article.
-
SIRT1 regulates Mxd1 during malignant melanoma progression.Oncotarget. 2017 Oct 3;8(70):114540-114553. doi: 10.18632/oncotarget.21457. eCollection 2017 Dec 29. Oncotarget. 2017. PMID: 29383100 Free PMC article.
-
Transcriptional signatures underlying dynamic phenotypic switching and novel disease biomarkers in a linear cellular model of melanoma progression.Neoplasia. 2021 Apr;23(4):439-455. doi: 10.1016/j.neo.2021.03.007. Epub 2021 Apr 9. Neoplasia. 2021. PMID: 33845354 Free PMC article.
-
Lysine Methyltransferase NSD1 and Cancers: Any Role in Melanoma?Cancers (Basel). 2022 Oct 5;14(19):4865. doi: 10.3390/cancers14194865. Cancers (Basel). 2022. PMID: 36230787 Free PMC article. Review.
-
Tackling malignant melanoma epigenetically: histone lysine methylation.Clin Epigenetics. 2018 Nov 22;10(1):145. doi: 10.1186/s13148-018-0583-z. Clin Epigenetics. 2018. PMID: 30466474 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

