Systematic analysis of small RNAs associated with human mitochondria by deep sequencing: detailed analysis of mitochondrial associated miRNA
- PMID: 22984580
- PMCID: PMC3439422
- DOI: 10.1371/journal.pone.0044873
Systematic analysis of small RNAs associated with human mitochondria by deep sequencing: detailed analysis of mitochondrial associated miRNA
Abstract
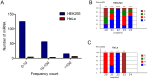
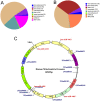
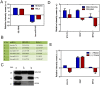
Mitochondria are one of the central regulators of many cellular processes beyond its well established role in energy metabolism. The inter-organellar crosstalk is critical for the optimal function of mitochondria. Many nuclear encoded proteins and RNA are imported to mitochondria. The translocation of small RNA (sRNA) including miRNA to mitochondria and other sub-cellular organelle is still not clear. We characterized here sRNA including miRNA associated with human mitochondria by cellular fractionation and deep sequencing approach. Mitochondria were purified from HEK293 and HeLa cells for RNA isolation. The sRNA library was generated and sequenced using Illumina system. The analysis showed the presence of unique population of sRNA associated with mitochondria including miRNA. Putative novel miRNAs were characterized from unannotated sRNA sequences. The study showed the association of 428 known, 196 putative novel miRNAs to mitochondria of HEK293 and 327 known, 13 putative novel miRNAs to mitochondria of HeLa cells. The alignment of sRNA to mitochondrial genome was also studied. The targets were analyzed using DAVID to classify them in unique networks using GO and KEGG tools. Analysis of identified targets showed that miRNA associated with mitochondria regulates critical cellular processes like RNA turnover, apoptosis, cell cycle and nucleotide metabolism. The six miRNAs (counts >1000) associated with mitochondria of both HEK293 and HeLa were validated by RT-qPCR. To our knowledge, this is the first systematic study demonstrating the associations of sRNA including miRNA with mitochondria that may regulate site-specific turnover of target mRNA important for mitochondrial related functions.
Conflict of interest statement
Figures
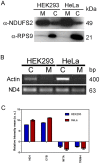
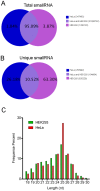
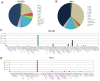
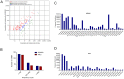
Similar articles
-
Nuclear outsourcing of RNA interference components to human mitochondria.PLoS One. 2011;6(6):e20746. doi: 10.1371/journal.pone.0020746. Epub 2011 Jun 13. PLoS One. 2011. PMID: 21695135 Free PMC article.
-
Identification of Taxus microRNAs and their targets with high-throughput sequencing and degradome analysis.Physiol Plant. 2012 Dec;146(4):388-403. doi: 10.1111/j.1399-3054.2012.01668.x. Epub 2012 Jul 25. Physiol Plant. 2012. PMID: 22708792
-
smRNAome profiling to identify conserved and novel microRNAs in Stevia rebaudiana Bertoni.BMC Plant Biol. 2012 Nov 1;12:197. doi: 10.1186/1471-2229-12-197. BMC Plant Biol. 2012. PMID: 23116282 Free PMC article.
-
The use of high-throughput sequencing methods for plant microRNA research.RNA Biol. 2015;12(7):709-19. doi: 10.1080/15476286.2015.1053686. RNA Biol. 2015. PMID: 26016494 Free PMC article. Review.
-
Interplay of mitochondrial metabolism and microRNAs.Cell Mol Life Sci. 2017 Feb;74(4):631-646. doi: 10.1007/s00018-016-2342-7. Epub 2016 Aug 25. Cell Mol Life Sci. 2017. PMID: 27563705 Free PMC article. Review.
Cited by
-
The roles of microRNA in redox metabolism and exercise-mediated adaptation.J Sport Health Sci. 2020 Sep;9(5):405-414. doi: 10.1016/j.jshs.2020.03.004. Epub 2020 Mar 19. J Sport Health Sci. 2020. PMID: 32780693 Free PMC article. Review.
-
mt tRFs, New Players in MELAS Disease.Front Physiol. 2022 Feb 22;13:800171. doi: 10.3389/fphys.2022.800171. eCollection 2022. Front Physiol. 2022. PMID: 35273517 Free PMC article.
-
MitosRNAs and extreme anoxia tolerance in embryos of the annual killifish Austrofundulus limnaeus.Sci Rep. 2019 Dec 24;9(1):19812. doi: 10.1038/s41598-019-56231-2. Sci Rep. 2019. PMID: 31874982 Free PMC article.
-
Changes in Mitochondrial Epigenome in Type 2 Diabetes Mellitus.Br J Biomed Sci. 2023 Feb 14;80:10884. doi: 10.3389/bjbs.2023.10884. eCollection 2023. Br J Biomed Sci. 2023. PMID: 36866104 Free PMC article. Review.
-
Mitochondrial Epigenetics Regulating Inflammation in Cancer and Aging.Front Cell Dev Biol. 2022 Jul 12;10:929708. doi: 10.3389/fcell.2022.929708. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35903542 Free PMC article. Review.
References
-
- Goetzman ES (2011) Modeling disorders of fatty acid metabolism in the mouse. Prog Mol Biol Transl Sci 100: 389–417. - PubMed
-
- Baltzer C, Tiefenbock SK, Frei C (2010) Mitochondria in response to nutrients and nutrient-sensitive pathways. Mitochondrion 10: 589–597. - PubMed
-
- Cloonan SM, Choi AM (2011) Mitochondria: commanders of innate immunity and disease? Current opinion in immunology. - PubMed
-
- Dimauro S, Garone C (2011) Metabolic disorders of fetal life: glycogenoses and mitochondrial defects of the mitochondrial respiratory chain. Seminars in fetal & neonatal medicine 16: 181–189. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

