Aging mechanism of soman inhibited acetylcholinesterase
- PMID: 22984913
- PMCID: PMC3475498
- DOI: 10.1021/jp307790v
Aging mechanism of soman inhibited acetylcholinesterase
Abstract
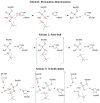
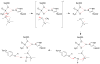
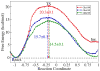
Acetylcholinesterase (AChE) is a crucial enzyme in the cholinergic nervous system that hydrolyzes neurotransmitter acetylcholine (ACh) and terminates synaptic signals. The catalytic serine of AChE can be phosphonylated by soman, one of the most potent nerve agents, and subsequently undergo an aging reaction. This phosphonylation and aging process leads to irreversible AChE inhibition, results in accumulation of excess ACh at the synaptic clefts, and causes neuromuscular paralysis. By employing Born-Oppenheimer ab initio QM/MM molecular dynamics simulations with umbrella sampling, a state-of-the-art approach to simulate enzyme reactions, we have characterized the aging mechanism of soman phosphonylated AChE and determined its free energy profile. This aging reaction starts with the scission of the O2-Cα bond, which is followed by methyl migration, and results in a tertiary carbenium intermediate. At the transition state, the scissile O2-Cα bond is already cleaved with an average O-C distance of 3.2 ± 0.3 Å and the migrating methyl group is shared between Cα and Cβ carbons with C-C distances of 1.9 ± 0.1 and 1.8 ± 0.1 Å, respectively. The negatively charged phosphonate group is stabilized by a salt bridge with the imidazole ring of the catalytic histidine. A major product of aging, 2,3-dimethyl-2-butanol can be formed swiftly by the reaction of a water molecule. Our characterized mechanism and simulation results provide new detailed insights into this important biochemical process.
Figures

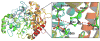



Similar articles
-
How is acetylcholinesterase phosphonylated by soman? An ab initio QM/MM molecular dynamics study.J Phys Chem A. 2014 Oct 2;118(39):9132-9. doi: 10.1021/jp502712d. Epub 2014 May 9. J Phys Chem A. 2014. PMID: 24786171 Free PMC article.
-
A first principles investigation of aging processes in soman conjugated AChE.Chem Biol Interact. 2013 Aug 25;204(3):185-90. doi: 10.1016/j.cbi.2013.05.013. Epub 2013 Jun 5. Chem Biol Interact. 2013. PMID: 23747845
-
Catalytic reaction mechanism of acetylcholinesterase determined by Born-Oppenheimer ab initio QM/MM molecular dynamics simulations.J Phys Chem B. 2010 Jul 8;114(26):8817-25. doi: 10.1021/jp104258d. J Phys Chem B. 2010. PMID: 20550161 Free PMC article.
-
Acetylcholinesterase inhibitors: a patent review (2008 - present).Expert Opin Ther Pat. 2012 Aug;22(8):871-86. doi: 10.1517/13543776.2012.701620. Epub 2012 Jul 6. Expert Opin Ther Pat. 2012. PMID: 22768972 Review.
-
Acetylcholinesterase inhibitors as Alzheimer therapy: from nerve toxins to neuroprotection.Eur J Med Chem. 2013;70:165-88. doi: 10.1016/j.ejmech.2013.09.050. Epub 2013 Oct 6. Eur J Med Chem. 2013. PMID: 24148993 Review.
Cited by
-
The recovery of acetylcholinesterase activity and the progression of neuropathological and pathophysiological alterations in the rat basolateral amygdala after soman-induced status epilepticus: relation to anxiety-like behavior.Neuropharmacology. 2014 Jun;81:64-74. doi: 10.1016/j.neuropharm.2014.01.035. Epub 2014 Jan 31. Neuropharmacology. 2014. PMID: 24486384 Free PMC article.
-
Computational Studies on Acetylcholinesterases.Molecules. 2017 Aug 10;22(8):1324. doi: 10.3390/molecules22081324. Molecules. 2017. PMID: 28796192 Free PMC article. Review.
-
A wrench in the works of human acetylcholinesterase: soman induced conformational changes revealed by molecular dynamics simulations.PLoS One. 2015 Apr 13;10(4):e0121092. doi: 10.1371/journal.pone.0121092. eCollection 2015. PLoS One. 2015. PMID: 25874456 Free PMC article.
-
Chemical, Physical, and Toxicological Properties of V-Agents.Int J Mol Sci. 2023 May 11;24(10):8600. doi: 10.3390/ijms24108600. Int J Mol Sci. 2023. PMID: 37239944 Free PMC article. Review.
-
A fluorogenic aryl fluorosulfate for intraorganellar transthyretin imaging in living cells and in Caenorhabditis elegans.J Am Chem Soc. 2015 Jun 17;137(23):7404-14. doi: 10.1021/jacs.5b03042. Epub 2015 Jun 8. J Am Chem Soc. 2015. PMID: 26051248 Free PMC article.
References
-
- Rosenberry TL. Adv Enzymol Relat Areas Mol Biol. John Wiley & Sons, Inc; 2006. pp. 103–218. - PubMed
-
- Quinn DM. Chem Rev. 1987;87:955–979.
-
- Silman I, Sussman JL. Curr Opin Pharmacol. 2005;5:293–302. - PubMed
-
- Sussman JL, Harel M, Frolow F, Oefner C, Goldman A, Toker L, Silman I. Science. 1991;253:872–879. - PubMed
-
- Goncalves AS, Franca TCC, Wilter A, Figueroa-Villar JD. J Braz Chem Soc. 2006;17:968–975.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

