Efficient experimental design and analysis strategies for the detection of differential expression using RNA-Sequencing
- PMID: 22985019
- PMCID: PMC3560154
- DOI: 10.1186/1471-2164-13-484
Efficient experimental design and analysis strategies for the detection of differential expression using RNA-Sequencing
Abstract
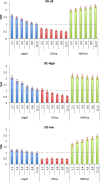
Background: RNA sequencing (RNA-Seq) has emerged as a powerful approach for the detection of differential gene expression with both high-throughput and high resolution capabilities possible depending upon the experimental design chosen. Multiplex experimental designs are now readily available, these can be utilised to increase the numbers of samples or replicates profiled at the cost of decreased sequencing depth generated per sample. These strategies impact on the power of the approach to accurately identify differential expression. This study presents a detailed analysis of the power to detect differential expression in a range of scenarios including simulated null and differential expression distributions with varying numbers of biological or technical replicates, sequencing depths and analysis methods.
Results: Differential and non-differential expression datasets were simulated using a combination of negative binomial and exponential distributions derived from real RNA-Seq data. These datasets were used to evaluate the performance of three commonly used differential expression analysis algorithms and to quantify the changes in power with respect to true and false positive rates when simulating variations in sequencing depth, biological replication and multiplex experimental design choices.
Conclusions: This work quantitatively explores comparisons between contemporary analysis tools and experimental design choices for the detection of differential expression using RNA-Seq. We found that the DESeq algorithm performs more conservatively than edgeR and NBPSeq. With regard to testing of various experimental designs, this work strongly suggests that greater power is gained through the use of biological replicates relative to library (technical) replicates and sequencing depth. Strikingly, sequencing depth could be reduced as low as 15% without substantial impacts on false positive or true positive rates.
Figures



References
-
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. - DOI - PMC - PubMed
-
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, Rinn JL, Lander ES, Regev A. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

