Clinical isolates of Yersinia enterocolitica biotype 1A represent two phylogenetic lineages with differing pathogenicity-related properties
- PMID: 22985268
- PMCID: PMC3512526
- DOI: 10.1186/1471-2180-12-208
Clinical isolates of Yersinia enterocolitica biotype 1A represent two phylogenetic lineages with differing pathogenicity-related properties
Abstract
Background: Y. enterocolitica biotype (BT) 1A strains are often isolated from human clinical samples but their contribution to disease has remained a controversial topic. Variation and the population structure among the clinical Y. enterocolitica BT 1A isolates have been poorly characterized. We used multi-locus sequence typing (MLST), 16S rRNA gene sequencing, PCR for ystA and ystB, lipopolysaccharide analysis, phage typing, human serum complement killing assay and analysis of the symptoms of the patients to characterize 298 clinical Y. enterocolitica BT 1A isolates in order to evaluate their relatedness and pathogenic potential.
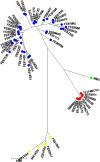
Results: A subset of 71 BT 1A strains, selected based on their varying LPS patterns, were subjected to detailed genetic analyses. The MLST on seven house-keeping genes (adk, argA, aroA, glnA, gyrB, thrA, trpE) conducted on 43 of the strains discriminated them into 39 MLST-types. By Bayesian analysis of the population structure (BAPS) the strains clustered conclusively into two distinct lineages, i.e. Genetic groups 1 and 2. The strains of Genetic group 1 were more closely related (97% similarity) to the pathogenic bio/serotype 4/O:3 strains than Genetic group 2 strains (95% similarity). Further comparison of the 16S rRNA genes of the BT 1A strains indicated that altogether 17 of the 71 strains belong to Genetic group 2. On the 16S rRNA analysis, these 17 strains were only 98% similar to the previously identified subspecies of Y. enterocolitica. The strains of Genetic group 2 were uniform in their pathogenecity-related properties: they lacked the ystB gene, belonged to the same LPS subtype or were of rough type, were all resistant to the five tested yersiniophages, were largely resistant to serum complement and did not ferment fucose. The 54 strains in Genetic group 1 showed much more variation in these properties. The most commonly detected LPS types were similar to the LPS types of reference strains with serotypes O:6,30 and O:6,31 (37%), O:7,8 (19%) and O:5 (15%).
Conclusions: The results of the present study strengthen the assertion that strains classified as Y. enterocolitica BT 1A represent more than one subspecies. Especially the BT 1A strains in our Genetic group 2 commonly showed resistance to human serum complement killing, which may indicate pathogenic potential for these strains. However, their virulence mechanisms remain unknown.
Figures

References
-
- Morris JG Jr, Prado V, Ferreccio C, Robins-Browne RM, Bordun AM, Cayazzo M, Kay BA, Levine MM. Yersinia enterocolitica isolated from two cohorts of young children in Santiago, Chile: incidence of and lack of correlation between illness and proposed virulence factors. J Clin Microbiol. 1991;29:2784–2788. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

