Single cell analysis of Vibrio harveyi uncovers functional heterogeneity in response to quorum sensing signals
- PMID: 22985329
- PMCID: PMC3511230
- DOI: 10.1186/1471-2180-12-209
Single cell analysis of Vibrio harveyi uncovers functional heterogeneity in response to quorum sensing signals
Abstract
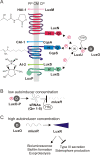
Background: Vibrio harveyi and closely related species are important pathogens in aquaculture. A complex quorum sensing cascade involving three autoinducers controls bioluminescence and several genes encoding virulence factors. Single cell analysis of a V. harveyi population has already indicated intercellular heterogeneity in the production of bioluminescence. This study was undertaken to analyze the expression of various autoinducer-dependent genes in individual cells.
Results: Here we used reporter strains bearing promoter::gfp fusions to monitor the induction/repression of three autoinducer-regulated genes in wild type conjugates at the single cell level. Two genes involved in pathogenesis - vhp and vscP, which code for an exoprotease and a component of the type III secretion system, respectively, and luxC (the first gene in the lux operon) were chosen for analysis. The lux operon and the exoprotease gene are induced, while vscP is repressed at high cell density. As controls luxS and recA, whose expression is not dependent on autoinducers, were examined. The responses of the promoter::gfp fusions in individual cells from the same culture ranged from no to high induction. Importantly, simultaneous analysis of two autoinducer induced phenotypes, bioluminescence (light detection) and exoproteolytic activity (fluorescence of a promoter::gfp fusion), in single cells provided evidence for functional heterogeneity within a V. harveyi population.
Conclusions: Autoinducers are not only an indicator for cell density, but play a pivotal role in the coordination of physiological activities within the population.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

