Identification of differentially expressed proteins in direct expressed prostatic secretions of men with organ-confined versus extracapsular prostate cancer
- PMID: 22986220
- PMCID: PMC3518113
- DOI: 10.1074/mcp.M112.017889
Identification of differentially expressed proteins in direct expressed prostatic secretions of men with organ-confined versus extracapsular prostate cancer
Abstract
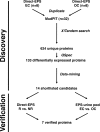
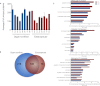
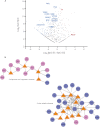
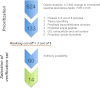
Current protocols for the screening of prostate cancer cannot accurately discriminate clinically indolent tumors from more aggressive ones. One reliable indicator of outcome has been the determination of organ-confined versus nonorgan-confined disease but even this determination is often only made following prostatectomy. This underscores the need to explore alternate avenues to enhance outcome prediction of prostate cancer patients. Fluids that are proximal to the prostate, such as expressed prostatic secretions (EPS), are attractive sources of potential prostate cancer biomarkers as these fluids likely bathe the tumor. Direct-EPS samples from 16 individuals with extracapsular (n = 8) or organ-confined (n = 8) prostate cancer were used as a discovery cohort, and were analyzed in duplicate by a nine-step MudPIT on a LTQ-Orbitrap XL mass spectrometer. A total of 624 unique proteins were identified by at least two unique peptides with a 0.2% false discovery rate. A semiquantitative spectral counting algorithm identified 133 significantly differentially expressed proteins in the discovery cohort. Integrative data mining prioritized 14 candidates, including two known prostate cancer biomarkers: prostate-specific antigen and prostatic acid phosphatase, which were significantly elevated in the direct-EPS from the organ-confined cancer group. These and five other candidates (SFN, MME, PARK7, TIMP1, and TGM4) were verified by Western blotting in an independent set of direct-EPS from patients with biochemically recurrent disease (n = 5) versus patients with no evidence of recurrence upon follow-up (n = 10). Lastly, we performed proof-of-concept SRM-MS-based relative quantification of the five candidates using unpurified heavy isotope-labeled synthetic peptides spiked into pools of EPS-urines from men with extracapsular and organ-confined prostate tumors. This study represents the first efforts to define the direct-EPS proteome from two major subclasses of prostate cancer using shotgun proteomics and verification in EPS-urine by SRM-MS.
Figures


Similar articles
-
Identification of prostate-enriched proteins by in-depth proteomic analyses of expressed prostatic secretions in urine.J Proteome Res. 2012 Apr 6;11(4):2386-96. doi: 10.1021/pr2011236. Epub 2012 Feb 29. J Proteome Res. 2012. PMID: 22339264 Free PMC article.
-
In-depth proteomic analyses of direct expressed prostatic secretions.J Proteome Res. 2010 May 7;9(5):2109-16. doi: 10.1021/pr1001498. J Proteome Res. 2010. PMID: 20334419 Free PMC article.
-
Clinical collection and protein properties of expressed prostatic secretions as a source for biomarkers of prostatic disease.J Proteomics. 2009 Aug 20;72(6):907-17. doi: 10.1016/j.jprot.2009.01.007. Epub 2009 Jan 20. J Proteomics. 2009. PMID: 19457353 Free PMC article. Review.
-
Multi-omics Biomarker Pipeline Reveals Elevated Levels of Protein-glutamine Gamma-glutamyltransferase 4 in Seminal Plasma of Prostate Cancer Patients.Mol Cell Proteomics. 2019 Sep;18(9):1807-1823. doi: 10.1074/mcp.RA119.001612. Epub 2019 Jun 27. Mol Cell Proteomics. 2019. PMID: 31249104 Free PMC article.
-
Proteomics in diagnosis of prostate cancer.Pril (Makedon Akad Nauk Umet Odd Med Nauki). 2015;36(1):5-36. Pril (Makedon Akad Nauk Umet Odd Med Nauki). 2015. PMID: 26076772 Review.
Cited by
-
Targeted glycoprotein enrichment and identification in stromal cell secretomes using azido sugar metabolic labeling.Proteomics Clin Appl. 2013 Jun;7(5-6):367-71. doi: 10.1002/prca.201300006. Proteomics Clin Appl. 2013. PMID: 23687070 Free PMC article.
-
The Latest Developments in Using Proteomic Biomarkers from Urine and Serum for Non-Invasive Disease Diagnosis and Prognosis.Biomark Insights. 2023 Jul 29;18:11772719231190218. doi: 10.1177/11772719231190218. eCollection 2023. Biomark Insights. 2023. PMID: 37528936 Free PMC article. Review.
-
Seminal plasma as a diagnostic fluid for male reproductive system disorders.Nat Rev Urol. 2014 May;11(5):278-88. doi: 10.1038/nrurol.2014.74. Epub 2014 Apr 8. Nat Rev Urol. 2014. PMID: 24709963 Review.
-
Systems pharmacology using mass spectrometry identifies critical response nodes in prostate cancer.NPJ Syst Biol Appl. 2018 Jul 2;4:26. doi: 10.1038/s41540-018-0064-1. eCollection 2018. NPJ Syst Biol Appl. 2018. PMID: 29977602 Free PMC article.
-
A novel expressed prostatic secretion (EPS)-urine metabolomic signature for the diagnosis of clinically significant prostate cancer.Cancer Biol Med. 2021 May 26;18(2):604-15. doi: 10.20892/j.issn.2095-3941.2020.0617. Cancer Biol Med. 2021. PMID: 34037347 Free PMC article.
References
-
- Greene K. L., Cowan J. E., Cooperberg M. R., Meng M. V., DuChane J., Carroll P. R. (2005) Who is the average patient presenting with prostate cancer? Urology 66, 76–82 - PubMed
-
- Ploussard G., de la Taille A. (2010) Urine biomarkers in prostate cancer. Nat. Rev. Urol. 7, 101–109 - PubMed
-
- Schröder F. H., Hugosson J., Roobol M. J., Tammela T. L., Ciatto S., Nelen V., Kwiatkowski M., Lujan M., Lilja H., Zappa M., Denis L. J., Recker F., Berenguer A., Määttänen L., Bangma C. H., Aus G., Villers A., Rebillard X., van der Kwast T., Blijenberg B. G., Moss S. M., de Koning H. J., Auvinen A. (2009) Screening and prostate-cancer mortality in a randomized European study. N. Engl. J. Med. 360, 1320–1328 - PubMed
-
- Freedland S. J., Humphreys E. B., Mangold L. A., Eisenberger M., Dorey F. J., Walsh P. C., Partin A. W. (2005) Risk of prostate cancer-specific mortality following biochemical recurrence after radical prostatectomy. JAMA 294, 433–439 - PubMed
-
- Penney K. L., Sinnott J. A., Fall K., Pawitan Y., Hoshida Y., Kraft P., Stark J. R., Fiorentino M., Perner S., Finn S., Calza S., Flavin R., Freedman M. L., Setlur S., Sesso H. D., Andersson S. O., Martin N., Kantoff P. W., Johansson J. E., Adami H. O., Rubin M. A., Loda M., Golub T. R., Andrén O., Stampfer M. J., Mucci L. A. (2011) mRNA expression signature of Gleason grade predicts lethal prostate cancer. J. Clin. Oncol. 29, 2391–2396 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

