CING: an integrated residue-based structure validation program suite
- PMID: 22986687
- PMCID: PMC3483101
- DOI: 10.1007/s10858-012-9669-7
CING: an integrated residue-based structure validation program suite
Abstract
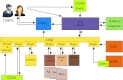
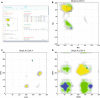
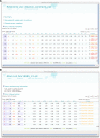
We present a suite of programs, named CING for Common Interface for NMR Structure Generation that provides for a residue-based, integrated validation of the structural NMR ensemble in conjunction with the experimental restraints and other input data. External validation programs and new internal validation routines compare the NMR-derived models with empirical data, measured chemical shifts, distance- and dihedral restraints and the results are visualized in a dynamic Web 2.0 report. A red-orange-green score is used for residues and restraints to direct the user to those critiques that warrant further investigation. Overall green scores below ~20 % accompanied by red scores over ~50 % are strongly indicative of poorly modelled structures. The publically accessible, secure iCing webserver ( https://nmr.le.ac.uk ) allows individual users to upload the NMR data and run a CING validation analysis.
Figures


Similar articles
-
NRG-CING: integrated validation reports of remediated experimental biomolecular NMR data and coordinates in wwPDB.Nucleic Acids Res. 2012 Jan;40(Database issue):D519-24. doi: 10.1093/nar/gkr1134. Epub 2011 Dec 1. Nucleic Acids Res. 2012. PMID: 22139937 Free PMC article.
-
An overview of tools for the validation of protein NMR structures.J Biomol NMR. 2014 Apr;58(4):259-85. doi: 10.1007/s10858-013-9750-x. Epub 2013 Jul 23. J Biomol NMR. 2014. PMID: 23877928 Review.
-
NMRe: a web server for NMR protein structure refinement with high-quality structure validation scores.Bioinformatics. 2016 Feb 15;32(4):611-3. doi: 10.1093/bioinformatics/btv595. Epub 2015 Oct 26. Bioinformatics. 2016. PMID: 26504145
-
GeNMR: a web server for rapid NMR-based protein structure determination.Nucleic Acids Res. 2009 Jul;37(Web Server issue):W670-7. doi: 10.1093/nar/gkp280. Epub 2009 Apr 30. Nucleic Acids Res. 2009. PMID: 19406927 Free PMC article.
-
Integrating Non-NMR Distance Restraints to Augment NMR Depiction of Protein Structure and Dynamics.J Mol Biol. 2020 Apr 17;432(9):2913-2929. doi: 10.1016/j.jmb.2020.01.023. Epub 2020 Feb 7. J Mol Biol. 2020. PMID: 32044345 Review.
Cited by
-
Backbone structure of Yersinia pestis Ail determined in micelles by NMR-restrained simulated annealing with implicit membrane solvation.J Biomol NMR. 2015 Sep;63(1):59-65. doi: 10.1007/s10858-015-9963-2. Epub 2015 Jul 5. J Biomol NMR. 2015. PMID: 26143069 Free PMC article.
-
Solution structure of the autophagy-related protein LC3C reveals a polyproline II motif on a mobile tether with phosphorylation site.Sci Rep. 2019 Oct 2;9(1):14167. doi: 10.1038/s41598-019-48155-8. Sci Rep. 2019. PMID: 31578424 Free PMC article.
-
Quality assessment of protein NMR structures.Curr Opin Struct Biol. 2013 Oct;23(5):715-24. doi: 10.1016/j.sbi.2013.08.005. Epub 2013 Sep 21. Curr Opin Struct Biol. 2013. PMID: 24060334 Free PMC article. Review.
-
Structure, dynamics and RNA binding of the multi-domain splicing factor TIA-1.Nucleic Acids Res. 2014 May;42(9):5949-66. doi: 10.1093/nar/gku193. Epub 2014 Mar 25. Nucleic Acids Res. 2014. PMID: 24682828 Free PMC article.
-
Structural Basis of Ca2+-Dependent Self-Processing Activity of Repeat-in-Toxin Proteins.mBio. 2020 Mar 17;11(2):e00226-20. doi: 10.1128/mBio.00226-20. mBio. 2020. PMID: 32184239 Free PMC article.
References
-
- Beck K, Andres C. Extreme programming explained: embrace change. 2. Boston: Addison-Wesley; 2004.
-
- Behnel S, Bradshaw R, Citro C, Dalcin L, Seljebotn DS, Smith K. Cython: the best of both worlds. Comput Sci Eng. 2011;13(2):31–39. doi: 10.1109/MCSE.2010.118. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

