Sonic hedgehog acts cell-autonomously on muscle precursor cells to generate limb muscle diversity
- PMID: 22987640
- PMCID: PMC3444735
- DOI: 10.1101/gad.187807.112
Sonic hedgehog acts cell-autonomously on muscle precursor cells to generate limb muscle diversity
Abstract
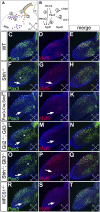
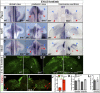
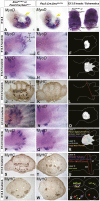
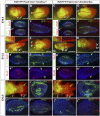
How muscle diversity is generated in the vertebrate body is poorly understood. In the limb, dorsal and ventral muscle masses constitute the first myogenic diversification, as each gives rise to distinct muscles. Myogenesis initiates after muscle precursor cells (MPCs) have migrated from the somites to the limb bud and populated the prospective muscle masses. Here, we show that Sonic hedgehog (Shh) from the zone of polarizing activity (ZPA) drives myogenesis specifically within the ventral muscle mass. Shh directly induces ventral MPCs to initiate Myf5 transcription and myogenesis through essential Gli-binding sites located in the Myf5 limb enhancer. In the absence of Shh signaling, myogenesis is delayed, MPCs fail to migrate distally, and ventral paw muscles fail to form. Thus, Shh production in the limb ZPA is essential for the spatiotemporal control of myogenesis and coordinates muscle and skeletal development by acting directly to regulate the formation of specific ventral muscles.
Figures


References
-
- Ahn S, Joyner AL 2004. Dynamic changes in the response of cells to positive hedgehog signaling during mouse limb patterning. Cell 118: 505–516 - PubMed
-
- Ahn K, Mishina Y, Hanks MC, Behringer RR, Crenshaw EB 3rd 2001. BMPR-IA signaling is required for the formation of the apical ectodermal ridge and dorsal–ventral patterning of the limb. Development 128: 4449–4461 - PubMed
-
- Amthor H, Christ B, Weil M, Patel K 1998. The importance of timing differentiation during limb muscle development. Curr Biol 8: 642–652 - PubMed
-
- Anderson C, Winder SJ, Borycki AG 2007. Dystroglycan protein distribution coincides with basement membranes and muscle differentiation during mouse embryogenesis. Dev Dyn 236: 2627–2635 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
