Engineering a model protein cavity to catalyze the Kemp elimination
- PMID: 22988064
- PMCID: PMC3479533
- DOI: 10.1073/pnas.1208076109
Engineering a model protein cavity to catalyze the Kemp elimination
Abstract
Synthetic cavitands and protein cavities have been widely studied as models for ligand recognition. Here we investigate the Met102 → His substitution in the artificial L99A cavity in T4 lysozyme as a Kemp eliminase. The resulting enzyme had k(cat)/K(M) = 0.43 M(-1) s(-1) and a (k(cat)/K(M))/k(uncat) = 10(7) at pH 5.0. The crystal structure of this enzyme was determined at 1.30 Å, as were the structures of four complexes of substrate and product analogs. The absence of ordered waters or hydrogen bonding interactions, and the presence of a common catalytic base (His102) in an otherwise hydrophobic, buried cavity, facilitated detailed analysis of the reaction mechanism and its optimization. Subsequent substitutions increased eliminase activity by an additional four-fold. As activity-enhancing substitutions were engineered into the cavity, protein stability decreased, consistent with the stability-function trade-off hypothesis. This and related model cavities may provide templates for studying protein design principles in radically simplified environments.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
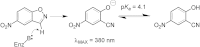


References
-
- Chang CE, Gilson MK. Free energy, entropy, and induced fit in host-guest recognition: Calculations with the second-generation mining minima algorithm. J Am Chem Soc. 2004;126:13156–13164. - PubMed
-
- Cram DJ, Cram JM. Cyclophane chemistry—Bent and battered benzene rings. Acc Chem Res. 1971;4:204–213.
-
- Eriksson AE, Baase WA, Wozniak JA, Matthews BW. A cavity-containing mutant of T4 lysozyme is stabilized by buried benzene. Nature. 1992;355:371–373. - PubMed
-
- Wei BQQ, Baase WA, Weaver LH, Matthews BW, Shoichet BK. A model binding site for testing scoring functions in molecular docking. J Mol Biol. 2002;322:339–355. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

