Complexity of chromatin folding is captured by the strings and binders switch model
- PMID: 22988072
- PMCID: PMC3479593
- DOI: 10.1073/pnas.1204799109
Complexity of chromatin folding is captured by the strings and binders switch model
Abstract
Chromatin has a complex spatial organization in the cell nucleus that serves vital functional purposes. A variety of chromatin folding conformations has been detected by single-cell imaging and chromosome conformation capture-based approaches. However, a unified quantitative framework describing spatial chromatin organization is still lacking. Here, we explore the "strings and binders switch" model to explain the origin and variety of chromatin behaviors that coexist and dynamically change within living cells. This simple polymer model recapitulates the scaling properties of chromatin folding reported experimentally in different cellular systems, the fractal state of chromatin, the processes of domain formation, and looping out. Additionally, the strings and binders switch model reproduces the recently proposed "fractal-globule" model, but only as one of many possible transient conformations.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

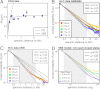

 ] is plotted as a function of s. It provides a measure of the relative amplitude of fluctuations of the polymer conformations. K = 1.50 when R2(s) is randomly distributed as a self-avoiding polymer (horizontal dashed line). Experimental K values depart from 1.50; K values were first analyzed in ref. , and originate from human fibroblast chromosome 1 ridges or whole chromosomes 1 or 11 (9) (squares, open circles, and filled circles, respectively), and from pre/pro-B or pro-B cell murine immunoglobulin heavy chain locus (10) (light- or dark-blue diamonds, respectively). (B) The kurtosis measured in the SBS model is plotted as a function of cm. K is close to 1.5 at low and high concentrations of binding molecules (open and closed chromatin). Around the binder threshold concentration, K exhibits a peak with values up to approximately 5. The range of values of K measured experimentally (A) matches the range found within the SBS model. It emerges that, beyond open and compact states, chromatin loci are likely to include also fractal conformations corresponding to the transition point.
] is plotted as a function of s. It provides a measure of the relative amplitude of fluctuations of the polymer conformations. K = 1.50 when R2(s) is randomly distributed as a self-avoiding polymer (horizontal dashed line). Experimental K values depart from 1.50; K values were first analyzed in ref. , and originate from human fibroblast chromosome 1 ridges or whole chromosomes 1 or 11 (9) (squares, open circles, and filled circles, respectively), and from pre/pro-B or pro-B cell murine immunoglobulin heavy chain locus (10) (light- or dark-blue diamonds, respectively). (B) The kurtosis measured in the SBS model is plotted as a function of cm. K is close to 1.5 at low and high concentrations of binding molecules (open and closed chromatin). Around the binder threshold concentration, K exhibits a peak with values up to approximately 5. The range of values of K measured experimentally (A) matches the range found within the SBS model. It emerges that, beyond open and compact states, chromatin loci are likely to include also fractal conformations corresponding to the transition point.
References
-
- Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: Regulation of gene expression in three dimensions. Nat Rev Genet. 2007;8:104–115. - PubMed
-
- Misteli T. Beyond the sequence: Cellular organization of genome function. Cell. 2007;128:787–800. - PubMed
-
- Cook PR. The organization of replication and transcription. Science. 1999;284:1790–1795. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

