Structural basis for the sheddase function of human meprin β metalloproteinase at the plasma membrane
- PMID: 22988105
- PMCID: PMC3479590
- DOI: 10.1073/pnas.1211076109
Structural basis for the sheddase function of human meprin β metalloproteinase at the plasma membrane
Abstract
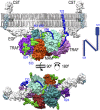
Ectodomain shedding at the cell surface is a major mechanism to regulate the extracellular and circulatory concentration or the activities of signaling proteins at the plasma membrane. Human meprin β is a 145-kDa disulfide-linked homodimeric multidomain type-I membrane metallopeptidase that sheds membrane-bound cytokines and growth factors, thereby contributing to inflammatory diseases, angiogenesis, and tumor progression. In addition, it cleaves amyloid precursor protein (APP) at the β-secretase site, giving rise to amyloidogenic peptides. We have solved the X-ray crystal structure of a major fragment of the meprin β ectoprotein, the first of a multidomain oligomeric transmembrane sheddase, and of its zymogen. The meprin β dimer displays a compact shape, whose catalytic domain undergoes major rearrangement upon activation, and reveals an exosite and a sugar-rich channel, both of which possibly engage in substrate binding. A plausible structure-derived working mechanism suggests that substrates such as APP are shed close to the plasma membrane surface following an "N-like" chain trace.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
The crystal structure of a 250-kDa heterotetrameric particle explains inhibition of sheddase meprin β by endogenous fetuin-B.Proc Natl Acad Sci U S A. 2021 Apr 6;118(14):e2023839118. doi: 10.1073/pnas.2023839118. Proc Natl Acad Sci U S A. 2021. PMID: 33782129 Free PMC article.
-
The metalloproteases meprin α and meprin β: unique enzymes in inflammation, neurodegeneration, cancer and fibrosis.Biochem J. 2013 Mar 1;450(2):253-64. doi: 10.1042/BJ20121751. Biochem J. 2013. PMID: 23410038 Free PMC article. Review.
-
Metalloprotease meprin β is activated by transmembrane serine protease matriptase-2 at the cell surface thereby enhancing APP shedding.Biochem J. 2015 Aug 15;470(1):91-103. doi: 10.1042/BJ20141417. Epub 2015 Jun 15. Biochem J. 2015. PMID: 26251449
-
Intersubunit and domain interactions of the meprin B metalloproteinase. Disulfide bonds and protein-protein interactions in the MAM and TRAF domains.J Biol Chem. 2005 Apr 8;280(14):13895-901. doi: 10.1074/jbc.M414218200. Epub 2005 Feb 4. J Biol Chem. 2005. PMID: 15695509
-
The Metalloprotease Meprin β Is an Alternative β-Secretase of APP.Front Mol Neurosci. 2017 Jan 5;9:159. doi: 10.3389/fnmol.2016.00159. eCollection 2016. Front Mol Neurosci. 2017. PMID: 28105004 Free PMC article. Review.
Cited by
-
The crystal structure of a 250-kDa heterotetrameric particle explains inhibition of sheddase meprin β by endogenous fetuin-B.Proc Natl Acad Sci U S A. 2021 Apr 6;118(14):e2023839118. doi: 10.1073/pnas.2023839118. Proc Natl Acad Sci U S A. 2021. PMID: 33782129 Free PMC article.
-
Characterization of the Cancer-Associated Meprin Βeta Variants G45R and G89R.Front Mol Biosci. 2021 Oct 6;8:702341. doi: 10.3389/fmolb.2021.702341. eCollection 2021. Front Mol Biosci. 2021. PMID: 34692768 Free PMC article.
-
The metalloproteases meprin α and meprin β: unique enzymes in inflammation, neurodegeneration, cancer and fibrosis.Biochem J. 2013 Mar 1;450(2):253-64. doi: 10.1042/BJ20121751. Biochem J. 2013. PMID: 23410038 Free PMC article. Review.
-
Hidden Relationships between N-Glycosylation and Disulfide Bonds in Individual Proteins.Int J Mol Sci. 2022 Mar 29;23(7):3742. doi: 10.3390/ijms23073742. Int J Mol Sci. 2022. PMID: 35409101 Free PMC article. Review.
-
Crystal structure of TRAF1 TRAF domain and its implications in the TRAF1-mediated intracellular signaling pathway.Sci Rep. 2016 May 6;6:25526. doi: 10.1038/srep25526. Sci Rep. 2016. PMID: 27151821 Free PMC article.
References
-
- Blobel CP. ADAMs: Key components in EGFR signalling and development. Nat Rev Mol Cell Biol. 2005;6:32–43. - PubMed
-
- Arribas J, Merlos-Suárez A. Shedding of plasma membrane proteins. Curr Top Dev Biol. 2003;54:125–144. - PubMed
-
- Pandiella A, Bosenberg MW, Huang EJ, Besmer P, Massagué J. Cleavage of membrane-anchored growth factors involves distinct protease activities regulated through common mechanisms. J Biol Chem. 1992;267:24028–24033. - PubMed
-
- Saftig P, Reiss K. The “A Disintegrin And Metalloproteases” ADAM10 and ADAM17: Novel drug targets with therapeutic potential? Eur J Cell Biol. 2011;90:527–535. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

