Regulation of bacterial photosynthesis genes by the small noncoding RNA PcrZ
- PMID: 22988125
- PMCID: PMC3479615
- DOI: 10.1073/pnas.1207067109
Regulation of bacterial photosynthesis genes by the small noncoding RNA PcrZ
Abstract
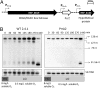
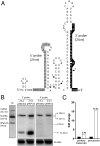
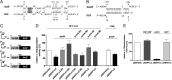
The small RNA PcrZ (photosynthesis control RNA Z) of the facultative phototrophic bacterium Rhodobacter sphaeroides is induced upon a drop of oxygen tension with similar kinetics to those of genes for components of photosynthetic complexes. High expression of PcrZ depends on PrrA, the response regulator of the PrrB/PrrA two-component system with a central role in redox regulation in R. sphaeroides. In addition the FnrL protein, an activator of some photosynthesis genes at low oxygen tension, is involved in redox-dependent expression of this small (s)RNA. Overexpression of full-length PcrZ in R. sphaeroides affects expression of a small subset of genes, most of them with a function in photosynthesis. Some mRNAs from the photosynthetic gene cluster were predicted to be putative PcrZ targets and results from an in vivo reporter system support these predictions. Our data reveal a negative effect of PcrZ on expression of its target mRNAs. Thus, PcrZ counteracts the redox-dependent induction of photosynthesis genes, which is mediated by protein regulators. Because PrrA directly activates photosynthesis genes and at the same time PcrZ, which negatively affects photosynthesis gene expression, this is one of the rare cases of an incoherent feed-forward loop including an sRNA. Our data identified PcrZ as a trans acting sRNA with a direct regulatory function in formation of photosynthetic complexes and provide a model for the control of photosynthesis gene expression by a regulatory network consisting of proteins and a small noncoding RNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Moskvin OV, Kaplan S, Gilles-Gonzalez MA, Gomelsky M. Novel heme-based oxygen sensor with a revealing evolutionary history. J Biol Chem. 2007;282:28740–28748. - PubMed
-
- Masuda S, Bauer CE. AppA is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell. 2002;110:613–623. - PubMed
-
- Braatsch S, Gomelsky M, Kuphal S, Klug G. A single flavoprotein, AppA, integrates both redox and light signals in Rhodobacter sphaeroides. Mol Microbiol. 2002;45:827–836. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

