Diverse developmental disorders from the one ring: distinct molecular pathways underlie the cohesinopathies
- PMID: 22988450
- PMCID: PMC3439829
- DOI: 10.3389/fgene.2012.00171
Diverse developmental disorders from the one ring: distinct molecular pathways underlie the cohesinopathies
Abstract
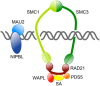
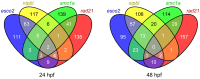
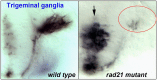
The multi-subunit protein complex, cohesin, is responsible for sister chromatid cohesion during cell division. The interaction of cohesin with DNA is controlled by a number of additional regulatory proteins. Mutations in cohesin, or its regulators, cause a spectrum of human developmental syndromes known as the "cohesinopathies." Cohesinopathy disorders include Cornelia de Lange Syndrome and Roberts Syndrome. The discovery of novel roles for chromatid cohesion proteins in regulating gene expression led to the idea that cohesinopathies are caused by dysregulation of multiple genes downstream of mutations in cohesion proteins. Consistent with this idea, Drosophila, mouse, and zebrafish cohesinopathy models all show altered expression of developmental genes. However, there appears to be incomplete overlap among dysregulated genes downstream of mutations in different components of the cohesion apparatus. This is surprising because mutations in all cohesion proteins would be predicted to affect cohesin's roles in cell division and gene expression in similar ways. Here we review the differences and similarities between genetic pathways downstream of components of the cohesion apparatus, and discuss how such differences might arise, and contribute to the spectrum of cohesinopathy disorders. We propose that mutations in different elements of the cohesion apparatus have distinct developmental outcomes that can be explained by sometimes subtly different molecular effects.
Keywords: CdLS; RBS; animal models; cohesin; gene expression regulation.
Figures

References
-
- Atienza J. M., Roth R. B., Rosette C., Smylie K. J., Kammerer S., Rehbock J., Ekblom J., Denissenko M. F. (2005). Suppression of RAD21 gene expression decreases cell growth and enhances cytotoxicity of etoposide and bleomycin in human breast cancer cells. Mol. Cancer Ther. 4, 361–368 - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

