CLCA2, a target of the p53 family, negatively regulates cancer cell migration and invasion
- PMID: 22990203
- PMCID: PMC3542243
- DOI: 10.4161/cbt.22280
CLCA2, a target of the p53 family, negatively regulates cancer cell migration and invasion
Abstract
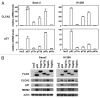
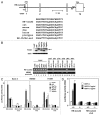
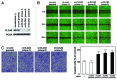
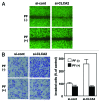
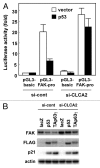
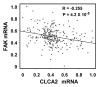
The tumor suppressor p53 transcriptionally regulates a number of genes that are involved in cell-cycle inhibition, apoptosis and the maintenance of genetic stability. Recent studies suggest that p53 also contributes to the regulation of cell migration and invasion. Here, we show that human chloride channel accessory-2 (CLCA2) is a target gene of the p53 family (p53, p73 and p63). CLCA2 is induced by DNA damage in a p53-dependent manner. The p53 family proteins activate the CLCA2 promoter by binding directly to the conserved consensus p53-binding site present in the CLCA2 promoter. In terms of function, ectopic expression of CLCA2 inhibited cancer cell migration. In contrast, silencing CLCA2 with siRNA stimulated cancer cell migration and invasion. We also found that inactivation of CLCA2 enhanced the expression of focal adhesion kinase (FAK), as well as its promoter activation. A small-molecule FAK inhibitor reduced the effect of CLCA2 siRNA on cell migration and invasion, suggesting that CLCA2 inhibits cancer cell migration and invasion through suppression of the FAK signaling pathway. Furthermore, there was an inverse correlation between CLCA2 and FAK expression in 251 human breast cancer tissues. These results strongly suggest that CLCA2 is involved in the p53 tumor suppressor network and has a significant effect on cell migration and invasion.
Figures

Similar articles
-
Along with its favorable prognostic role, CLCA2 inhibits growth and metastasis of nasopharyngeal carcinoma cells via inhibition of FAK/ERK signaling.J Exp Clin Cancer Res. 2018 Feb 20;37(1):34. doi: 10.1186/s13046-018-0692-8. J Exp Clin Cancer Res. 2018. PMID: 29463274 Free PMC article.
-
Identification and characterization of the intercellular adhesion molecule-2 gene as a novel p53 target.Oncotarget. 2016 Sep 20;7(38):61426-61437. doi: 10.18632/oncotarget.11366. Oncotarget. 2016. PMID: 27556181 Free PMC article.
-
CLCA2 as a p53-inducible senescence mediator.Neoplasia. 2012 Feb;14(2):141-9. doi: 10.1593/neo.111700. Neoplasia. 2012. PMID: 22431922 Free PMC article.
-
Focal adhesion kinase and p53 signal transduction pathways in cancer.Front Biosci (Landmark Ed). 2010 Jun 1;15(3):901-12. doi: 10.2741/3653. Front Biosci (Landmark Ed). 2010. PMID: 20515733 Free PMC article. Review.
-
FAK and p53 protein interactions.Anticancer Agents Med Chem. 2011 Sep;11(7):617-9. doi: 10.2174/187152011796817619. Anticancer Agents Med Chem. 2011. PMID: 21355845 Free PMC article. Review.
Cited by
-
CLCA2 as a novel immunohistochemical marker for differential diagnosis of squamous cell carcinoma from adenocarcinoma of the lung.Dis Markers. 2014;2014:619273. doi: 10.1155/2014/619273. Epub 2014 Dec 7. Dis Markers. 2014. PMID: 25548429 Free PMC article.
-
CLCA2 overexpression suppresses epithelial-to-mesenchymal transition in cervical cancer cells through inactivation of ERK/JNK/p38-MAPK signaling pathways.BMC Mol Cell Biol. 2022 Oct 25;23(1):44. doi: 10.1186/s12860-022-00440-7. BMC Mol Cell Biol. 2022. PMID: 36280802 Free PMC article.
-
BluePrint breast cancer molecular subtyping recognizes single and dual subtype tumors with implications for therapeutic guidance.Breast Cancer Res Treat. 2022 Oct;195(3):263-274. doi: 10.1007/s10549-022-06698-x. Epub 2022 Aug 19. Breast Cancer Res Treat. 2022. PMID: 35984580 Free PMC article.
-
RANK-RANKL Signaling in Cancer of the Uterine Cervix: A Review.Int J Mol Sci. 2019 May 2;20(9):2183. doi: 10.3390/ijms20092183. Int J Mol Sci. 2019. PMID: 31052546 Free PMC article. Review.
-
<em>miR-19a</em> targeting <em>CLCA4</em> to regulate the proliferation, migration, and invasion of colorectal cancer cells.Eur J Histochem. 2022 Mar 10;66(1):3381. doi: 10.4081/ejh.2022.3381. Eur J Histochem. 2022. PMID: 35266369 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
