Hrp3 controls nucleosome positioning to suppress non-coding transcription in eu- and heterochromatin
- PMID: 22990236
- PMCID: PMC3512395
- DOI: 10.1038/emboj.2012.267
Hrp3 controls nucleosome positioning to suppress non-coding transcription in eu- and heterochromatin
Abstract
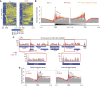
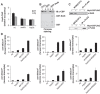
The positioning of the nucleosome by ATP-dependent remodellers provides the fundamental chromatin environment for the regulation of diverse cellular processes acting on the underlying DNA. Recently, genome-wide nucleosome mapping has revealed more detailed information on the chromatin-remodelling factors. Here, we report that the Schizosaccharomyces pombe CHD remodeller, Hrp3, is a global regulator that drives proper nucleosome positioning and nucleosome stability. The loss of Hrp3 resulted in nucleosome perturbation across the chromosome, and the production of antisense transcripts in the hrp3Δ cells emphasized the importance of nucleosome architecture for proper transcription. Notably, perturbation of the nucleosome in hrp3 deletion mutant was also associated with destabilization of the DNA-histone interaction and cell cycle-dependent alleviation of heterochromatin silencing. Furthermore, the effect of Hrp3 in the pericentric region was found to be accomplished via a physical interaction with Swi6, and appeared to cooperate with other heterochromatin factors for gene silencing. Taken together, our data indicate that a well-positioned nucleosome by Hrp3 is important for the spatial-temporal control of transcription-associated processes.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





Comment in
-
Nucleosome positioning and transcription: fission yeast CHD remodellers make their move.EMBO J. 2012 Nov 28;31(23):4371-2. doi: 10.1038/emboj.2012.284. Epub 2012 Oct 26. EMBO J. 2012. PMID: 23103764 Free PMC article.
Similar articles
-
Nucleosome positioning and transcription: fission yeast CHD remodellers make their move.EMBO J. 2012 Nov 28;31(23):4371-2. doi: 10.1038/emboj.2012.284. Epub 2012 Oct 26. EMBO J. 2012. PMID: 23103764 Free PMC article.
-
The Role of Non-Catalytic Domains of Hrp3 in Nucleosome Remodeling.Int J Mol Sci. 2021 Feb 11;22(4):1793. doi: 10.3390/ijms22041793. Int J Mol Sci. 2021. PMID: 33670267 Free PMC article.
-
CHD1 remodelers regulate nucleosome spacing in vitro and align nucleosomal arrays over gene coding regions in S. pombe.EMBO J. 2012 Nov 28;31(23):4388-403. doi: 10.1038/emboj.2012.289. Epub 2012 Oct 26. EMBO J. 2012. PMID: 23103765 Free PMC article.
-
Opposing Roles of FACT for Euchromatin and Heterochromatin in Yeast.Biomolecules. 2023 Feb 16;13(2):377. doi: 10.3390/biom13020377. Biomolecules. 2023. PMID: 36830746 Free PMC article. Review.
-
Destabilizing heterochromatin: Does Swi6/HP1 make the choice?Mol Cell. 2006 Jun 23;22(6):709-710. doi: 10.1016/j.molcel.2006.06.004. Mol Cell. 2006. PMID: 16793539 Review.
Cited by
-
Chd1 co-localizes with early transcription elongation factors independently of H3K36 methylation and releases stalled RNA polymerase II at introns.Epigenetics Chromatin. 2014 Oct 27;7(1):32. doi: 10.1186/1756-8935-7-32. eCollection 2014. Epigenetics Chromatin. 2014. PMID: 25395991 Free PMC article.
-
Nucleosomal signatures impose nucleosome positioning in coding and noncoding sequences in the genome.Genome Res. 2016 Nov;26(11):1532-1543. doi: 10.1101/gr.207241.116. Epub 2016 Sep 23. Genome Res. 2016. PMID: 27662899 Free PMC article.
-
Nucleosome positioning in yeasts: methods, maps, and mechanisms.Chromosoma. 2015 Jun;124(2):131-51. doi: 10.1007/s00412-014-0501-x. Epub 2014 Dec 23. Chromosoma. 2015. PMID: 25529773 Review.
-
Noncoding Transcription Is a Driving Force for Nucleosome Instability in spt16 Mutant Cells.Mol Cell Biol. 2016 Jun 15;36(13):1856-67. doi: 10.1128/MCB.00152-16. Print 2016 Jul 1. Mol Cell Biol. 2016. PMID: 27141053 Free PMC article.
-
The 19S proteasome is directly involved in the regulation of heterochromatin spreading in fission yeast.J Biol Chem. 2017 Oct 13;292(41):17144-17155. doi: 10.1074/jbc.M117.790824. Epub 2017 Aug 7. J Biol Chem. 2017. PMID: 28784663 Free PMC article.
References
-
- Bernardi F, Koller T, Thoma F (1991) The ade6 gene of the fission yeast Schizosaccharomyces pombe has the same chromatin structure in the chromosome and in plasmids. Yeast 7: 547–558 - PubMed
-
- Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SIS (2005) Comprehensive analysis of heterochromatin-and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet 37: 809–819 - PubMed
-
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia W-J, Anderson S, Yates J, Washburn MP, Workman JL (2005) Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

