AAV vectors containing rDNA homology display increased chromosomal integration and transgene persistence
- PMID: 22990673
- PMCID: PMC3464636
- DOI: 10.1038/mt.2012.157
AAV vectors containing rDNA homology display increased chromosomal integration and transgene persistence
Abstract
Although recombinant adeno-associated viral (rAAV) vectors are promising tools for gene therapy of genetic disorders, they remain mostly episomal and hence are lost during cell replication. For this reason, rAAV vectors capable of chromosomal integration would be desirable. Ribosomal DNA (rDNA) repeat sequences are overrepresented during random integration of rAAV. We therefore sought to enhance AAV integration frequency by including 28S rDNA homology arms into our vector design. A vector containing ~1 kb of homology on each side of a cDNA expression cassette for human fumarylacetoacetate hydrolase (FAH) was constructed. rAAV of serotypes 2 and 8 were injected into Fah-deficient mice, a model for human tyrosinemia type 1. Integrated FAH transgenes are positively selected in this model and rDNA-containing AAV vectors had a ~30× higher integration frequency than controls. Integration by homologous recombination (HR) into the 28S rDNA locus was seen in multiple tissues. Furthermore, rDNA-containing AAV vectors for human factor IX (hFIX) demonstrated increased transgene persistence after liver regeneration. We conclude that rDNA containing AAV vectors may be superior to conventional vector design for the treatment of genetic diseases, especially those associated with increased hepatocyte replication.
Figures



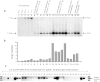

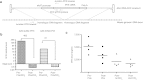
Comment in
-
AAV vectors for the nucleolus.Mol Ther. 2012 Oct;20(10):1842-3. doi: 10.1038/mt.2012.193. Mol Ther. 2012. PMID: 23023058 Free PMC article. No abstract available.
Similar articles
-
Ribosomal DNA integrating rAAV-rDNA vectors allow for stable transgene expression.Mol Ther. 2012 Oct;20(10):1912-23. doi: 10.1038/mt.2012.164. Epub 2012 Sep 18. Mol Ther. 2012. PMID: 22990671 Free PMC article.
-
Herpes simplex virus type 1/adeno-associated virus hybrid vectors mediate site-specific integration at the adeno-associated virus preintegration site, AAVS1, on human chromosome 19.J Virol. 2002 Jul;76(14):7163-73. doi: 10.1128/jvi.76.14.7163-7173.2002. J Virol. 2002. PMID: 12072516 Free PMC article.
-
Hybrid adeno-associated viral vectors utilizing transposase-mediated somatic integration for stable transgene expression in human cells.PLoS One. 2013 Oct 8;8(10):e76771. doi: 10.1371/journal.pone.0076771. eCollection 2013. PLoS One. 2013. PMID: 24116154 Free PMC article.
-
Expressing Transgenes That Exceed the Packaging Capacity of Adeno-Associated Virus Capsids.Hum Gene Ther Methods. 2016 Feb;27(1):1-12. doi: 10.1089/hgtb.2015.140. Hum Gene Ther Methods. 2016. PMID: 26757051 Free PMC article. Review.
-
[Integration of AAV vectors and insertional mutagenesis].Med Sci (Paris). 2016 Feb;32(2):167-74. doi: 10.1051/medsci/20163202010. Epub 2016 Mar 2. Med Sci (Paris). 2016. PMID: 26936174 Review. French.
Cited by
-
Viral Vectors in Gene Therapy: Where Do We Stand in 2023?Viruses. 2023 Mar 7;15(3):698. doi: 10.3390/v15030698. Viruses. 2023. PMID: 36992407 Free PMC article. Review.
-
Progress in Gene Therapy for Hereditary Tyrosinemia Type 1.Pharmaceutics. 2025 Mar 18;17(3):387. doi: 10.3390/pharmaceutics17030387. Pharmaceutics. 2025. PMID: 40143050 Free PMC article. Review.
-
rAAV-mediated tumorigenesis: still unresolved after an AAV assault.Mol Ther. 2012 Nov;20(11):2014-7. doi: 10.1038/mt.2012.220. Mol Ther. 2012. PMID: 23131853 Free PMC article. No abstract available.
-
A universal system to select gene-modified hepatocytes in vivo.Sci Transl Med. 2016 Jun 8;8(342):342ra79. doi: 10.1126/scitranslmed.aad8166. Sci Transl Med. 2016. PMID: 27280686 Free PMC article.
-
Emerging Issues in AAV-Mediated In Vivo Gene Therapy.Mol Ther Methods Clin Dev. 2017 Dec 1;8:87-104. doi: 10.1016/j.omtm.2017.11.007. eCollection 2018 Mar 16. Mol Ther Methods Clin Dev. 2017. PMID: 29326962 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

